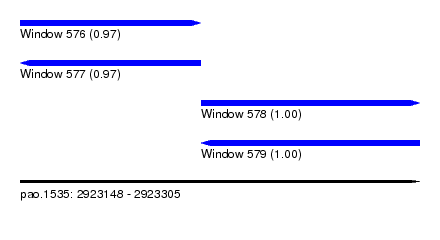
| Sequence ID | pao.1535 |
|---|---|
| Location | 2,923,148 – 2,923,305 |
| Length | 157 |
| Max. P | 0.999898 |

| Location | 2,923,148 – 2,923,219 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 53.94 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -23.00 |
| Energy contribution | -19.85 |
| Covariance contribution | -3.15 |
| Combinations/Pair | 1.96 |
| Mean z-score | -6.12 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1535 2923148 71 + 6264404 CGGAACGAAAAAAGGGCCCGAAGGCCCUUUCU-CAAUGAUCUACAGACUUCA--UUGGAACUCU-AUAGAUCUAU ......((..((((((((....)))))))).)-)...((((((.(((.(((.--...))).)))-.))))))... ( -23.90) >pau.1474 3878868 64 - 6537648 CGAAACGAAAAAAGGGCCCUAAGGCCCUUUUUUCAAUGAUCUACAGGC----------AAGCCU-GUGGAUCUAU ......((((((((((((....))))))))))))...((((((((((.----------...)))-)))))))... ( -36.40) >pss.1735 3473676 70 + 6093698 AGAAACGAAAAAAGACGCCUAGGCGUCUUUUU-UGCUGAUUCACAAAGCCCG---GAGGGUUCA-GUGAAUCAAU .......(((((((((((....))))))))))-)..((((((((..(((((.---..)))))..-)))))))).. ( -30.90) >pf5.2155 4107880 70 + 7074893 AGAAACGAAAAAAGGACCCGAAGGUCCUUUUU-UCGAUCGCUGCAGAAUUCG---UGGAACUCU-GCAGCGCUAU .....(((((((((((((....))))))))))-)))..(((((((((.(((.---..))).)))-)))))).... ( -36.00) >psp.1406 3222012 71 - 5928787 AAAAACGAAAAAAGACGCCUAAGCGUCUUUUU-UGUUAAUCCCAUAUGCCUC---UUGUGCAUACAUGGAUUUAU ...(((.(((((((((((....))))))))))-))))(((((..((((((..---..).)))))...)))))... ( -22.60) >pst.1796 3397686 63 + 6397126 -------AAAAAAGAGGCAUAAGCCUCUUUUU-C-UGGUUUCCACACACUUCAAUUGAAGCGCG-UUGAA--AAU -------.((((((((((....))))))))))-.-...((((.((.(.((((....)))).).)-).)))--).. ( -20.90) >consensus AGAAACGAAAAAAGAGCCCUAAGCCCCUUUUU_CAAUGAUCCACAAAACUCC___UGGAACUCU_GUGGAUCUAU ........((((((((((....)))))))))).....((((((((((.(((......))).))).)))))))... (-23.00 = -19.85 + -3.15)

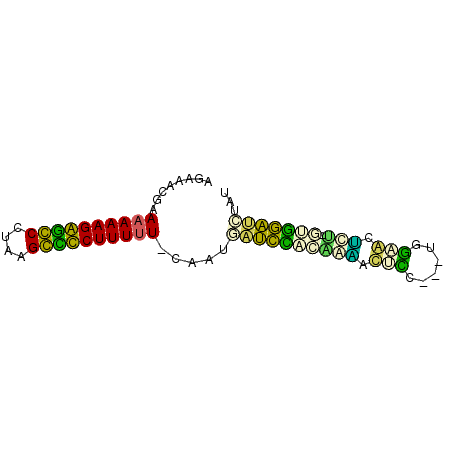
| Location | 2,923,148 – 2,923,219 |
|---|---|
| Length | 71 |
| Sequences | 6 |
| Columns | 75 |
| Reading direction | reverse |
| Mean pairwise identity | 53.94 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -29.50 |
| Energy contribution | -25.88 |
| Covariance contribution | -3.62 |
| Combinations/Pair | 2.04 |
| Mean z-score | -7.31 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1535 2923148 71 - 6264404 AUAGAUCUAU-AGAGUUCCAA--UGAAGUCUGUAGAUCAUUG-AGAAAGGGCCUUCGGGCCCUUUUUUCGUUCCG ...(((((((-(((.(((...--.))).))))))))))...(-((((((((((....)))))))))))....... ( -36.40) >pau.1474 3878868 64 + 6537648 AUAGAUCCAC-AGGCUU----------GCCUGUAGAUCAUUGAAAAAAGGGCCUUAGGGCCCUUUUUUCGUUUCG ...((((.((-(((...----------.))))).))))...((((((((((((....))))))))))))...... ( -33.40) >pss.1735 3473676 70 - 6093698 AUUGAUUCAC-UGAACCCUC---CGGGCUUUGUGAAUCAGCA-AAAAAGACGCCUAGGCGUCUUUUUUCGUUUCU ..((((((((-.((.(((..---.))).)).))))))))(((-((((((((((....))))))))))).)).... ( -28.70) >pf5.2155 4107880 70 - 7074893 AUAGCGCUGC-AGAGUUCCA---CGAAUUCUGCAGCGAUCGA-AAAAAGGACCUUCGGGUCCUUUUUUCGUUUCU ....((((((-(((((((..---.)))))))))))))..(((-((((((((((....)))))))))))))..... ( -43.40) >psp.1406 3222012 71 + 5928787 AUAAAUCCAUGUAUGCACAA---GAGGCAUAUGGGAUUAACA-AAAAAGACGCUUAGGCGUCUUUUUUCGUUUUU ...(((((.(((((((....---...))))))))))))((((-((((((((((....))))))))))).)))... ( -25.40) >pst.1796 3397686 63 - 6397126 AUU--UUCAA-CGCGCUUCAAUUGAAGUGUGUGGAAACCA-G-AAAAAGAGGCUUAUGCCUCUUUUUU------- ..(--(((.(-((((((((....))))))))).))))...-(-((((((((((....)))))))))))------- ( -29.30) >consensus AUAGAUCCAC_AGAGCUCCA___CGAGCUCUGUAGAUCAACA_AAAAAGAGCCUUAGGCCCCUUUUUUCGUUUCU ...(((((((.(((((((......)))))))))))))).....((((((((((....))))))))))........ (-29.50 = -25.88 + -3.62)

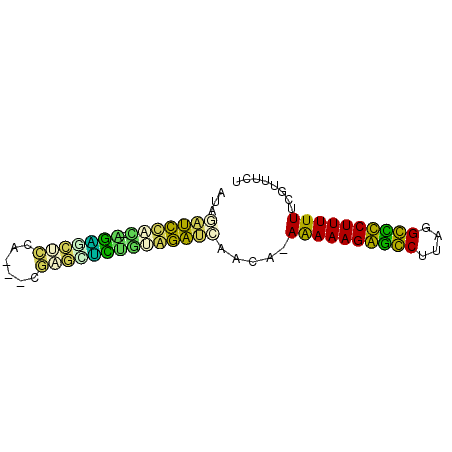
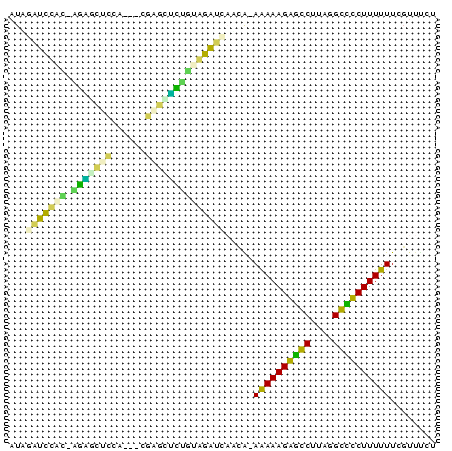
| Location | 2,923,219 – 2,923,305 |
|---|---|
| Length | 86 |
| Sequences | 6 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1535 2923219 86 + 6264404 AUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUCGUCU .....(((((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).)).... ( -30.40) >pau.1474 3878932 86 - 6537648 AUCUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUCGUCU .....(((((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).)).... ( -30.40) >pss.1735 3473746 83 + 6093698 GUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUG--- .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))....--- ( -29.90) >pf5.2155 4107950 83 + 7074893 GUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAACG--- ....(..(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))..).--- ( -30.00) >pfo.2462 4728948 83 + 6438405 AUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUG--- .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))....--- ( -29.90) >pst.1796 3397749 83 + 6397126 AUCUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUUG--- .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))....--- ( -29.90) >consensus AUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUCG___ .......(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))....... (-29.90 = -29.90 + 0.00)

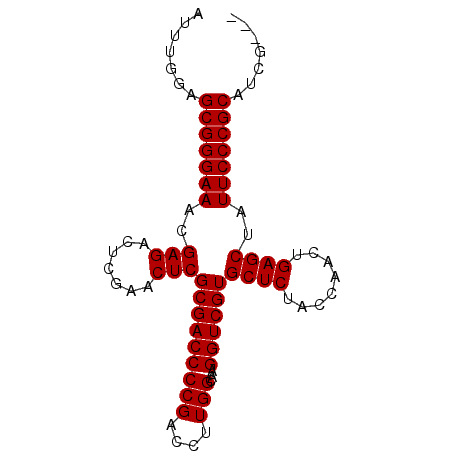
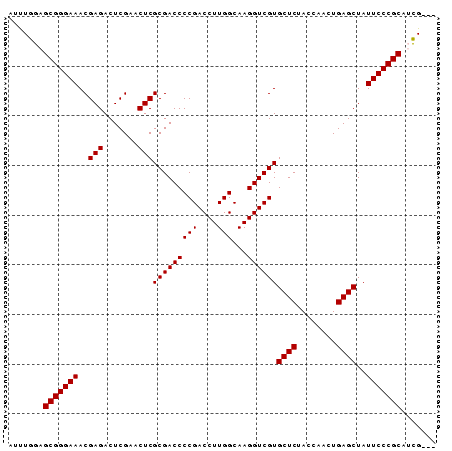
| Location | 2,923,219 – 2,923,305 |
|---|---|
| Length | 86 |
| Sequences | 6 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.44 |
| SVM RNA-class probability | 0.999898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1535 2923219 86 - 6264404 AGACGAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAU ....((.(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))))..... ( -34.20) >pau.1474 3878932 86 + 6537648 AGACGAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGAU ....((.(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))))..... ( -34.20) >pss.1735 3473746 83 - 6093698 ---CAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAC ---....(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))....... ( -33.60) >pf5.2155 4107950 83 - 7074893 ---CGUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAC ---.(..(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))..).... ( -34.30) >pfo.2462 4728948 83 - 6438405 ---CAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAU ---....(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))....... ( -33.60) >pst.1796 3397749 83 - 6397126 ---CAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAGAU ---....(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))....... ( -33.60) >consensus ___CAAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAU .......(((((((..((((........))))..((((((((....).)))))))(((((.......))))))))))))....... (-33.60 = -33.60 + 0.00)

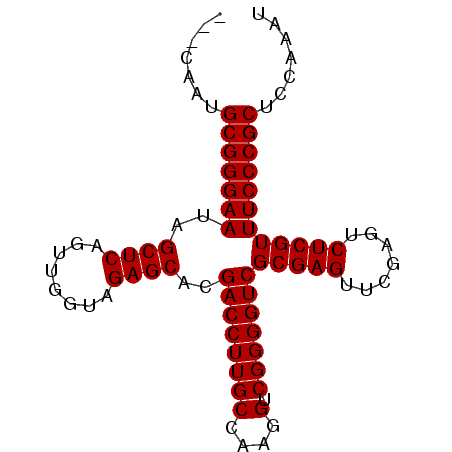
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:44 2007