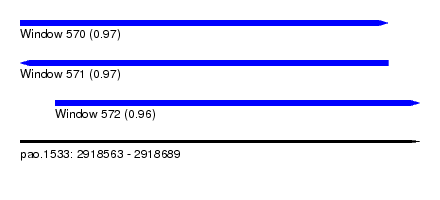
| Sequence ID | pao.1533 |
|---|---|
| Location | 2,918,563 – 2,918,689 |
| Length | 126 |
| Max. P | 0.968268 |

| Location | 2,918,563 – 2,918,679 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -50.88 |
| Consensus MFE | -36.95 |
| Energy contribution | -37.78 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1533 2918563 116 + 6264404 AAAAAGCCCCGUAACUCAC-UGAGCUACGGGG-CUUUCC-UGUUGGAGGCUGAGGUCGGAAUCGAACCGGCGUUCACGGAUUUGCAAUCCGGUGCAUAACCACUCUGCUACUCAGCCUU ..(((((((((((.(((..-.))).)))))))-))))..-.....((((((((((((((.......)))))((...(((((.....)))))..))...............))))))))) ( -50.40) >pau.1477 3872503 116 - 6537648 AAAAAGCCCCGUAACUCAC-UGAGCUACGGGG-CUUUCC-UGUUGGAGGCUGAGGUCGGAAUCGAACCGGCGUUCACGGAUUUGCAAUCCGGUGCAUAACCACUCUGCUACUCAGCCUU ..(((((((((((.(((..-.))).)))))))-))))..-.....((((((((((((((.......)))))((...(((((.....)))))..))...............))))))))) ( -50.40) >pel.1299 3520912 117 - 5888780 GAAAAACCCCGCAGA-CCUGUGAUCUGCGGGG-UUUCGAAUGAUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGACGGAUUUGCAAUCCGGAGCAUAACCACUUUGCUACUCGGCCUC ...((((((((((((-.......)))))))))-))).........((((((((((((((.......))))).....(((((.....))))).((((.........)))).))))))))) ( -53.80) >ppk.2263 4427473 106 + 6181863 GAAAAACCCCGCAAC-CUCAAUGGCUGCGGGGCUUUCGAAUGGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGACGGAUUUGCAAUCCGGAGCAUAACCACUUUG------------ ((((..(((((((.(-(.....)).))))))).))))....(((((..((((.(.(((....))).)))))((...(((((.....)))))..))....)))))...------------ ( -41.80) >pf5.2093 3982007 116 + 7074893 AAAAAACCCCGUAGA-CGU-UAAUCUACGGGG-UUUUCAAGGGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGGCGGAUUUGCAAUCCGCUGCAUAACCAUUUUGCUACUCGGCCCC ..(((((((((((((-...-...)))))))))-)))).......((.((((((((((((.......)))))((((.(((((.....)))))))))...............))))))))) ( -54.40) >pst.1776 3351504 116 + 6397126 AAAAAACCCCGCAGA-CGU-UAAUCUGCGGGG-CUUUGAAUAGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGGCGGAUUUGCAAUCCGCUGCAUAACCAUUUUGCUACUCGGCCUC ..(((.(((((((((-...-...)))))))))-.)))........((((((((((((((.......)))))((((.(((((.....)))))))))...............))))))))) ( -54.50) >consensus AAAAAACCCCGCAAA_CAC_UGAGCUACGGGG_CUUUCAAUGGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGACGGAUUUGCAAUCCGGUGCAUAACCACUUUGCUACUCGGCCUC ..(((((((((((............))))))).))))........((((((((((((((.......)))))((...(((((.....)))))..))...............))))))))) (-36.95 = -37.78 + 0.83)

| Location | 2,918,563 – 2,918,679 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -51.38 |
| Consensus MFE | -39.11 |
| Energy contribution | -38.00 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.46 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1533 2918563 116 - 6264404 AAGGCUGAGUAGCAGAGUGGUUAUGCACCGGAUUGCAAAUCCGUGAACGCCGGUUCGAUUCCGACCUCAGCCUCCAACA-GGAAAG-CCCCGUAGCUCA-GUGAGUUACGGGGCUUUUU .((((((((...(.((((.(......(((((.(..(......)..)...))))).).)))).)..))))))))......-((((((-(((((((((((.-..))))))))))))))))) ( -52.00) >pau.1477 3872503 116 + 6537648 AAGGCUGAGUAGCAGAGUGGUUAUGCACCGGAUUGCAAAUCCGUGAACGCCGGUUCGAUUCCGACCUCAGCCUCCAACA-GGAAAG-CCCCGUAGCUCA-GUGAGUUACGGGGCUUUUU .((((((((...(.((((.(......(((((.(..(......)..)...))))).).)))).)..))))))))......-((((((-(((((((((((.-..))))))))))))))))) ( -52.00) >pel.1299 3520912 117 + 5888780 GAGGCCGAGUAGCAAAGUGGUUAUGCUCCGGAUUGCAAAUCCGUCUACGCCGGUUCGAUUCCGACCUCGGCCUCCAUCAUUCGAAA-CCCCGCAGAUCACAGG-UCUGCGGGGUUUUUC (((((((((..((..(((((...(((........)))...))).))..))(((.......)))..)))))))))........((((-((((((((((.....)-))))))))))))).. ( -53.60) >ppk.2263 4427473 106 - 6181863 ------------CAAAGUGGUUAUGCUCCGGAUUGCAAAUCCGUCUACGCCGGUUCGAUUCCGACCUCGGCCUCCACCAUUCGAAAGCCCCGCAGCCAUUGAG-GUUGCGGGGUUUUUC ------------...((((((.......(((((.....))))).....(((((.(((....))).).))))....)))))).(((((((((((((((.....)-)))))))))))))). ( -45.10) >pf5.2093 3982007 116 - 7074893 GGGGCCGAGUAGCAAAAUGGUUAUGCAGCGGAUUGCAAAUCCGCCUACGCCGGUUCGAUUCCGACCUCGGCCUCCACCCUUGAAAA-CCCCGUAGAUUA-ACG-UCUACGGGGUUUUUU ((((((((((((((.........))).((((((.....)))))).)))...(((.((....))))))))))).))......(((((-((((((((((..-..)-)))))))))))))). ( -52.00) >pst.1776 3351504 116 - 6397126 GAGGCCGAGUAGCAAAAUGGUUAUGCAGCGGAUUGCAAAUCCGCCUACGCCGGUUCGAUUCCGACCUCGGCCUCCACUAUUCAAAG-CCCCGCAGAUUA-ACG-UCUGCGGGGUUUUUU ((((((((((((((.........))).((((((.....)))))).)))...(((.((....)))))))))))))........((((-((((((((((..-..)-))))))))))))).. ( -53.60) >consensus GAGGCCGAGUAGCAAAGUGGUUAUGCACCGGAUUGCAAAUCCGUCUACGCCGGUUCGAUUCCGACCUCGGCCUCCACCAUUCAAAA_CCCCGCAGAUCA_ACG_GCUACGGGGUUUUUU .((((((((..(((.........)))..((((...(.((((.((....)).)))).)..))))..)))))))).........((((.(((((((((........))))))))))))).. (-39.11 = -38.00 + -1.11)

| Location | 2,918,574 – 2,918,689 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -30.70 |
| Energy contribution | -30.03 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1533 2918574 115 + 6264404 UAACUCAC-UGAGCUACGGGGCUU-UCC-UGUUGGAGGCUGAGGUCGGAAUCGAACCGGCGUUCACGGAUUUGCAAUCCGGUGCAUAACCACUCUGCUACUCAGCCUU-UGAGCGAAGC .......(-(..((..((((....-.))-))(..((((((((((((((.......)))))((...(((((.....)))))..))...............)))))))))-..)))..)). ( -39.90) >pau.1477 3872514 115 - 6537648 UAACUCAC-UGAGCUACGGGGCUU-UCC-UGUUGGAGGCUGAGGUCGGAAUCGAACCGGCGUUCACGGAUUUGCAAUCCGGUGCAUAACCACUCUGCUACUCAGCCUU-UGAGCGAAGC .......(-(..((..((((....-.))-))(..((((((((((((((.......)))))((...(((((.....)))))..))...............)))))))))-..)))..)). ( -39.90) >pel.1299 3520923 109 - 5888780 CAGA-CCUGUGAUCUGCGGGGUUU-CGAAUGAUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGACGGAUUUGCAAUCCGGAGCAUAACCACUUUGCUACUCGGCCUC-AAA------- ....-(((((.....)))))....-.........((((((((((((((.......))))).....(((((.....))))).((((.........)))).)))))))))-...------- ( -41.30) >pf5.2093 3982018 109 + 7074893 UAGA-CGU-UAAUCUACGGGGUUU-UCAAGGGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGGCGGAUUUGCAAUCCGCUGCAUAACCAUUUUGCUACUCGGCCCC-AAA------U ....-(((-......)))......-.......(((.((((((((((((.......)))))((((.(((((.....)))))))))...............)))))))))-)..------. ( -39.00) >pst.1776 3351515 109 + 6397126 CAGA-CGU-UAAUCUGCGGGGCUU-UGAAUAGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGGCGGAUUUGCAAUCCGCUGCAUAACCAUUUUGCUACUCGGCCUC-AUA------C ....-(((-......)))..(((.-.....))).((((((((((((((.......)))))((((.(((((.....)))))))))...............)))))))))-...------. ( -40.70) >pfo.1799 3506720 94 + 6438405 -------------------AGCUUGUUUAUAGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGGCGGAUUUGCAAUCCGCUGCAUAACCAUUUUGCUACUCGGCCUCAAAA------C -------------------...............((((((((((((((.......)))))((((.(((((.....)))))))))...............)))))))))....------. ( -38.00) >consensus UAGA_CAU_UGAUCUACGGGGCUU_UCAAUGGUGGAGGCCGAGGUCGGAAUCGAACCGGCGUAGACGGAUUUGCAAUCCGCUGCAUAACCACUUUGCUACUCGGCCUC_AAA______C ..................................((((((((((((((.......)))))((.(.(((((.....)))))).))...............)))))))))........... (-30.70 = -30.03 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:37 2007