
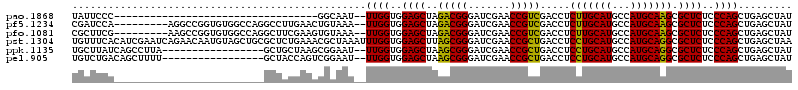
| Sequence ID | pao.1868 |
|---|---|
| Location | 3,514,659 – 3,515,218 |
| Length | 559 |
| Max. P | 0.999095 |

| Location | 3,514,659 – 3,514,775 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -47.09 |
| Consensus MFE | -41.05 |
| Energy contribution | -42.17 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.95 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
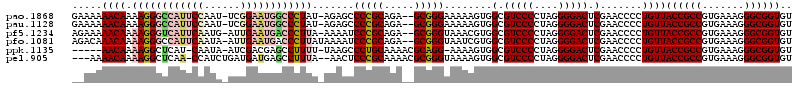
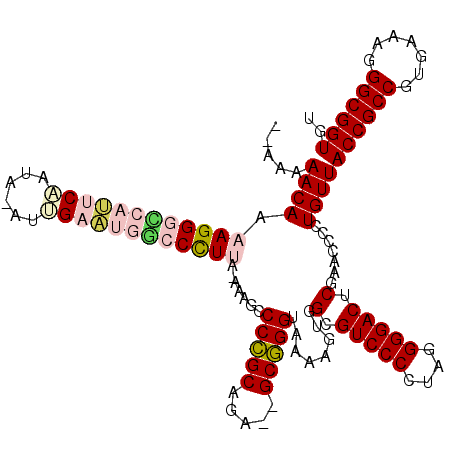
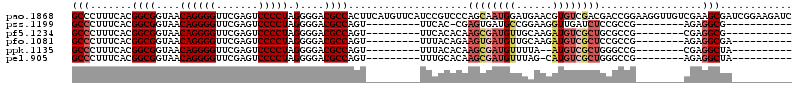
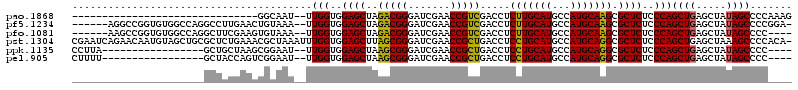
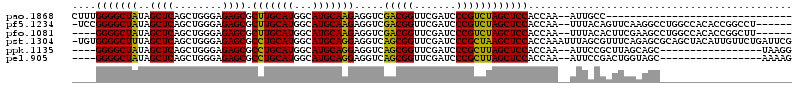
>pao.1868 3514659 116 + 6264404 GAAAAAACAAAAGGGCCAUUCCAAU-UCGGAAUGGCCCUAU-AGAGCCCCGCAGA--GCGGGAAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU ...........(((((((((((...-..)))))))))))((-((...(((((...--)))))........(.(((((....))))).)......))))(((((((.......))))))). ( -52.60) >pau.1128 4484020 116 - 6537648 GAAAAAACAAAAGGGCCAUUCCAAU-UCGGAAUGGCCCUAU-AGAGCCCCGCAGA--GCGGGAAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU ...........(((((((((((...-..)))))))))))((-((...(((((...--)))))........(.(((((....))))).)......))))(((((((.......))))))). ( -52.60) >pf5.1234 4794636 116 - 7074893 AGAAAAACAAAAGGGUCAUUCAAUG-AUUGAAUGACCCUUA-AAAAUCCCGCAGA--GCGGGUAAACGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU .....((((.((((((((((((...-..)))))))))))).-.....(((((...--)))))........(.(((((....))))).).......))))((((((.......)))))).. ( -47.40) >pfo.1081 4293631 117 - 6438405 AGACAAACAAAAGGGCCAUUCAAUA-AUUGAAUGACCCUUAUAAAAUCCCGCAGA--GCGGGUAAUCGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU .((((.....(((((.((((((...-..)))))).))))).......(((((...--)))))...(((....(((((....))))).))).....))))((((((.......)))))).. ( -43.90) >ppk.1135 3939805 111 - 6181863 -----AACAAAAGGCUCAU-CAAUA-AUCGACGAGCCUUUU-UAAGCCCUGCAAAACGCAGG-AAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU -----...(((((((((.(-(....-...)).)))))))))-.....(((((.....)))))-.......(.(((((....))))).)..........(((((((.......))))))). ( -42.80) >pel.905 4179519 114 - 5888780 ---AAAACAAAAGGCUCAA-CCAUCUGAUGAUGAGCCUUUA--AACUCCCGCAAAACGCGGGUAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU ---..((((...((.((..-.((((....)))).(((....--....(((((.....))))).......)))(((((....)))))..)).))..))))((((((.......)))))).. ( -43.26) >consensus __AAAAACAAAAGGGCCAUUCAAUA_AUUGAAUGGCCCUUA_AAAGCCCCGCAGA__GCGGGUAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGU .....((((.((((((((((((......)))))))))))).......(((((.....)))))........(.(((((....))))).).......))))((((((.......)))))).. (-41.05 = -42.17 + 1.12)
| Location | 3,514,659 – 3,514,775 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.88 |
| Mean single sequence MFE | -50.46 |
| Consensus MFE | -42.62 |
| Energy contribution | -43.32 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
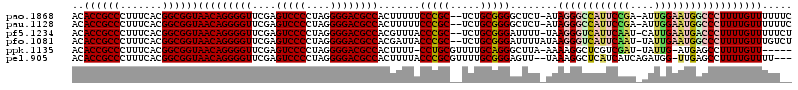
>pao.1868 3514659 116 - 6264404 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUUCCCGC--UCUGCGGGGCUCU-AUAGGGCCAUUCCGA-AUUGGAAUGGCCCUUUUGUUUUUUC ..((((((.......))))))(((((((((....(((((....)))))..........(((((--...))))))))))-..(((((((((((..-...)))))))))))..))))..... ( -54.10) >pau.1128 4484020 116 + 6537648 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUUCCCGC--UCUGCGGGGCUCU-AUAGGGCCAUUCCGA-AUUGGAAUGGCCCUUUUGUUUUUUC ..((((((.......))))))(((((((((....(((((....)))))..........(((((--...))))))))))-..(((((((((((..-...)))))))))))..))))..... ( -54.10) >pf5.1234 4794636 116 + 7074893 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACGUUUACCCGC--UCUGCGGGAUUUU-UAAGGGUCAUUCAAU-CAUUGAAUGACCCUUUUGUUUUUCU ..((((((.......))))))...((..(..((((((((....)))))..........(((((--...))))).....-.((((((((((((..-...)))))))))))))))..)..)) ( -49.70) >pfo.1081 4293631 117 + 6438405 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACGAUUACCCGC--UCUGCGGGAUUUUAUAAGGGUCAUUCAAU-UAUUGAAUGGCCCUUUUGUUUGUCU ..((((((.......))))))....(((..(((.(((((....)))))....)))...)))..--......((((..((.((((((((((((..-...)))))))))))).))...)))) ( -48.40) >ppk.1135 3939805 111 + 6181863 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUU-CCUGCGUUUUGCAGGGCUUA-AAAAGGCUCGUCGAU-UAUUG-AUGAGCCUUUUGUU----- ..((((((.......))))))...((((((....(((((....)))))))).))).(-(((((.....))))))....-((((((((((((((.-..)))-)))))))))))...----- ( -48.70) >pel.905 4179519 114 + 5888780 ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUACCCGCGUUUUGCGGGAGUU--UAAAGGCUCAUCAUCAGAUGG-UUGAGCCUUUUGUUUU--- ..((((((.......))))))((((((((((((((((((....)))))(((.......(((((.....)))))....--....)))(((((....)))))-))))))).))))))..--- ( -47.76) >consensus ACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUACCCGC__UCUGCGGGACUUU_AAAGGGCCAUUCAAU_AAUGGAAUGACCCUUUUGUUUUU__ ..((((((.......))))))(((((((((....(((((....)))))))).......(((((.....)))))........((((((((((((....))))))))))))))))))..... (-42.62 = -43.32 + 0.70)
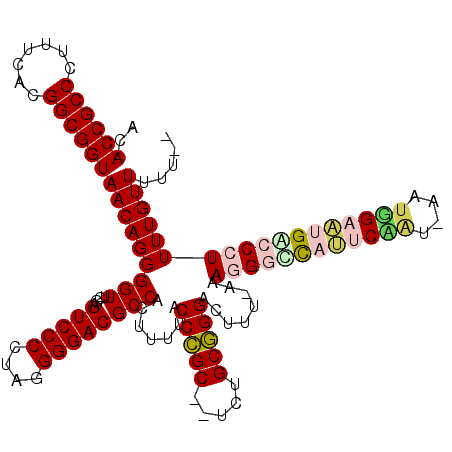
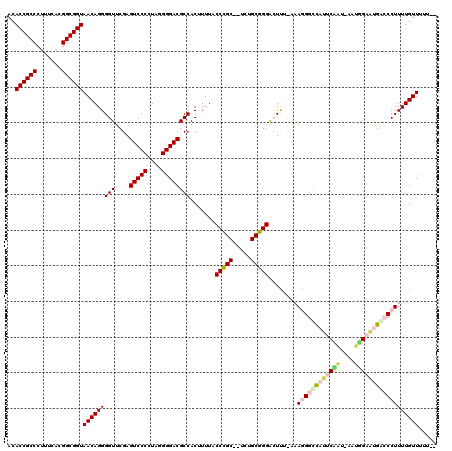
| Location | 3,514,698 – 3,514,815 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -49.37 |
| Consensus MFE | -45.99 |
| Energy contribution | -45.72 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514698 117 + 6264404 U-AGAGCCCCGCAGA--GCGGGAAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAAUGAAACCUUCGAAA .-.....(((((...--)))))....(((((((((((....)))))........(((.(((((((.......)))))))...)))))))))...((((((...........))))))... ( -49.90) >pau.1128 4484059 117 - 6537648 U-AGAGCCCCGCAGA--GCGGGAAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAAUGAAACCUUCGAAA .-.....(((((...--)))))....(((((((((((....)))))........(((.(((((((.......)))))))...)))))))))...((((((...........))))))... ( -49.90) >pf5.1234 4794675 116 - 7074893 A-AAAAUCCCGCAGA--GCGGGUAAACGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACA-CAAACCCUUCUAAC .-.....(((((...--))))).....((((((((((....)))))........(((.(((((((.......)))))))...)))))))).....((((((....-....)))))).... ( -49.90) >pfo.1081 4293670 117 - 6438405 AUAAAAUCCCGCAGA--GCGGGUAAUCGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACA-CAAACCUUCUAUAC .......(((((...--))))).....((((.(((((....))))).)...(((((...((((((.......))))))(((.(((....))))))..))))).))-)............. ( -46.10) >ppk.1135 3939838 111 - 6181863 U-UAAGCCCUGCAAAACGCAGG-AAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC---- .-.....(((((.....)))))-....((.((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))-).)--)....---- ( -49.80) >pel.905 4179555 111 - 5888780 A--AACUCCCGCAAAACGCGGGUAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC---- .--....(((((.....))))).....((.((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))-).)--)....---- ( -50.60) >consensus A_AAAGCCCCGCAGA__GCGGGUAAAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACA_UAAA_CCUUC_AA_ .......(((((.....)))))..........((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))................ (-45.99 = -45.72 + -0.28)

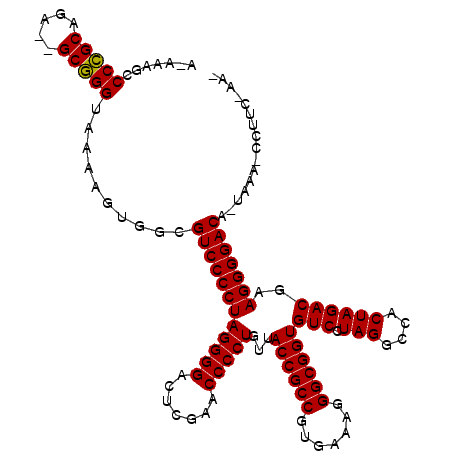
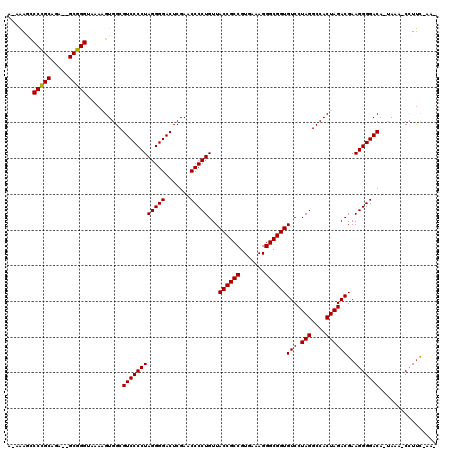
| Location | 3,514,698 – 3,514,815 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -47.77 |
| Consensus MFE | -44.13 |
| Energy contribution | -43.63 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514698 117 - 6264404 UUUCGAAGGUUUCAUUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUUCCCGC--UCUGCGGGGCUCU-A ...((((((...........))))))...((((((...((((((((((.......)))))).......))))..(((((....)))))))))))...((((((--...))))))....-. ( -45.81) >pau.1128 4484059 117 + 6537648 UUUCGAAGGUUUCAUUGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUUCCCGC--UCUGCGGGGCUCU-A ...((((((...........))))))...((((((...((((((((((.......)))))).......))))..(((((....)))))))))))...((((((--...))))))....-. ( -45.81) >pf5.1234 4794675 116 + 7074893 GUUAGAAGGGUUUG-UGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACGUUUACCCGC--UCUGCGGGAUUUU-U ....((((((....-....)))))).....(((((...((((((((((.......)))))).......))))..(((((....)))))))))).....(((((--...))))).....-. ( -48.81) >pfo.1081 4293670 117 + 6438405 GUAUAGAAGGUUUG-UGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACGAUUACCCGC--UCUGCGGGAUUUUAU ..((((((.((..(-((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))).)).....(((((--...))))).)))))) ( -47.30) >ppk.1135 3939838 111 + 6181863 ----GAAGG--UUA-CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUU-CCUGCGUUUUGCAGGGCUUA-A ----(((((--(..-((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))..))))))-(((((.....))))).....-. ( -48.30) >pel.905 4179555 111 + 5888780 ----GAAGG--UUA-CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUACCCGCGUUUUGCGGGAGUU--U ----(((((--(..-((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))..)))))).(((((.....)))))....--. ( -50.60) >consensus _UU_GAAGG_UUUA_UGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUUUACCCGC__UCUGCGGGACUUU_A ...............((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))).........(((((.....)))))....... (-44.13 = -43.63 + -0.50)

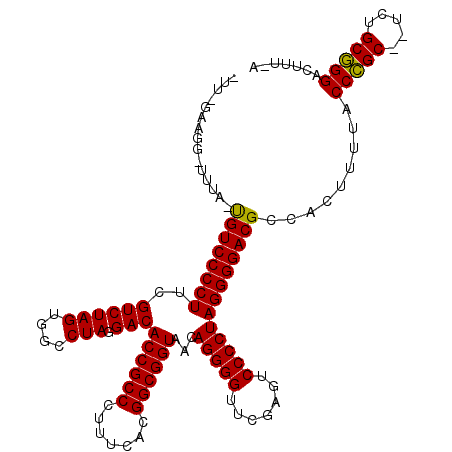
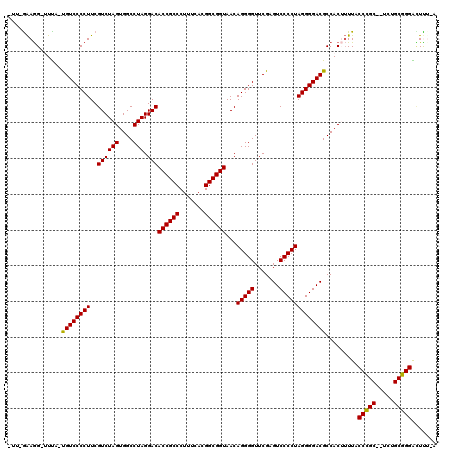
| Location | 3,514,735 – 3,514,832 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514735 97 + 6264404 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAAUGAAACCUUCGAAAAGUAAGGCC-AGCGAUGC---- ((((.......))))....((((((.......))))))(((((.(((((((...((((((...........))))))...)))..))))-)).)))..---- ( -36.30) >pau.1128 4484096 97 - 6537648 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACAAUGAAACCUUCGAAAAGUAAGGCC-AGCGAUGC---- ((((.......))))....((((((.......))))))(((((.(((((((...((((((...........))))))...)))..))))-)).)))..---- ( -36.30) >pss.1199 3796879 95 - 6093698 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-CAAAACCUUCAGUCAGGUUCAUCGAACGAU------ (....)..(((((((((.(((((((.......)))))))...))))......((((((((.....-.....))))).))).)))))..........------ ( -32.90) >pst.1304 4011899 94 - 6397126 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-CAAA-CCUUCAGUCAGGAUCACCGAGCGGU------ ((((.......))))....((((((.(....).)(((((((((((....)))((((((((.....-....-))))).))).))).))))).)))))------ ( -35.20) >ppk.1135 3939876 91 - 6181863 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC--------GUGCCGAUCCGCUAACGC ((((.......))))...(((((((.......)))))))..((((....))))(((((((.....-...--)))))--------))........((....)) ( -33.80) >pel.905 4179593 91 - 5888780 GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC--------GUGCCGAUCCGUUUUCAC ((((.......))))..........(((((.(((((((...((((....))))(((((((.....-...--)))))--------)))))).)))...))))) ( -33.80) >consensus GGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG_UAAA_CCUUC____AG_AACGCCGAGCGAU______ ((((.......))))....((((((.......))))))(((.(((....))))))(((((...........))))).......................... (-26.88 = -26.88 + 0.00)

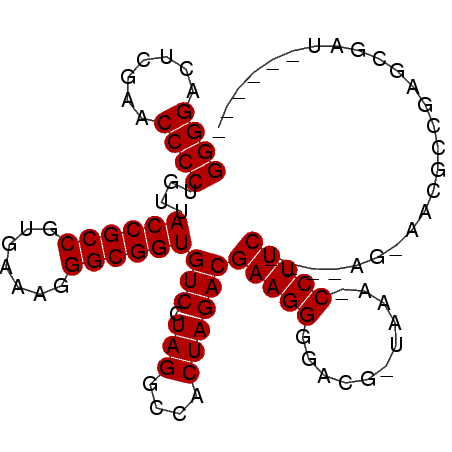
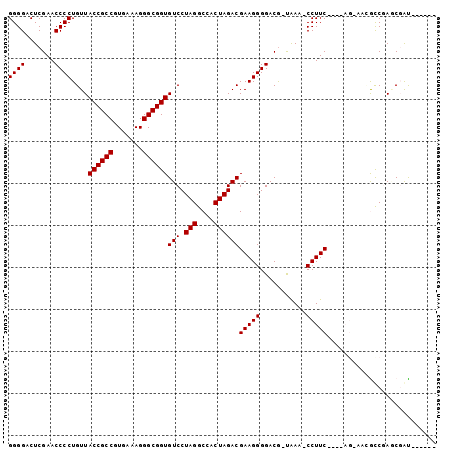
| Location | 3,514,775 – 3,514,861 |
|---|---|
| Length | 86 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -12.12 |
| Energy contribution | -12.07 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.991956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514775 86 + 6264404 CCUAGGCCACUAGACGAAGGGGACAAUGAAACCUUCGAAAAGUAAGGCC-AGCGAUGC----UGGCCUUG--GAAUGUGGUGGAGCUAGACGG .((((.((((((.(((((((...........)))))).....(((((((-(((...))----))))))))--...).))))))..)))).... ( -37.80) >psp.1169 3635875 86 - 5928787 CCUAGGCCACUAGACGAAGGGGACG-CAAAACCUUCAGUCAGGCUCAUCGAACGAU------GAACCCUGGCGAAUUUGGUGGAGCUUAGCGG ((((((((((((((.(((((.....-.....))))).((((((.(((((....)))------))..))))))...)))))))..)))))).). ( -33.90) >pss.1199 3796919 86 - 6093698 CCUAGGCCACUAGACGAAGGGGACG-CAAAACCUUCAGUCAGGUUCAUCGAACGAU------GAACCCUGGCAAAAUUGGUGGAGCUUAGCGG (((((((((((((..(((((.....-.....))))).((((((((((((....)))------))).))))))....))))))..)))))).). ( -35.20) >pst.1304 4011939 84 - 6397126 CCUAGGCCACUAGACGAAGGGGACG-CAAA-CCUUCAGUCAGGAUCACCGAGCGGU------GAACCCUGGCAAA-UUGGUGGAGCUUAGCGG (((((((((((((..(((((.....-....-))))).((((((.(((((....)))------))..))))))...-))))))..)))))).). ( -35.00) >ppk.1135 3939916 82 - 6181863 CCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC--------GUGCCGAUCCGCUAACGCGAACCGCGGCAAAAUUGGUGGAGCUAAGCGG ..(((.((((((((((((((.....-...--)))))--------))((((...(((....))).....))))....)))))))..)))..... ( -33.20) >pel.905 4179633 82 - 5888780 CCUAGGCCACUAGACGAAGGGGACG-UAA--CCUUC--------GUGCCGAUCCGUUUUCACGAACCGCGGCAAAAUUGGUGGAGCUAAGCGG ..(((.((((((((((((((.....-...--)))))--------))((((....((((....))))..))))....)))))))..)))..... ( -28.80) >consensus CCUAGGCCACUAGACGAAGGGGACG_CAAA_CCUUCAGUCAGG_UCACCGAACGAU______GAACCCUGGCAAAAUUGGUGGAGCUAAGCGG ...((.(((((((..(((((...........)))))..........(((....)))....................)))))))..))...... (-12.12 = -12.07 + -0.06)

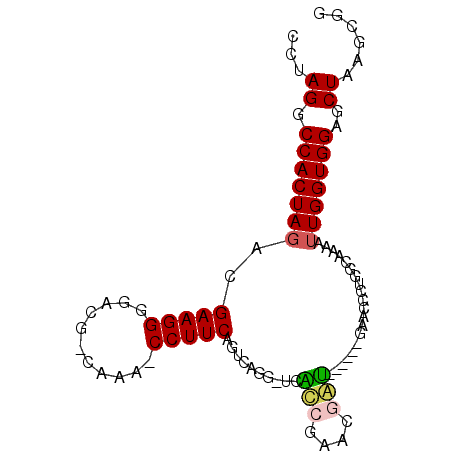
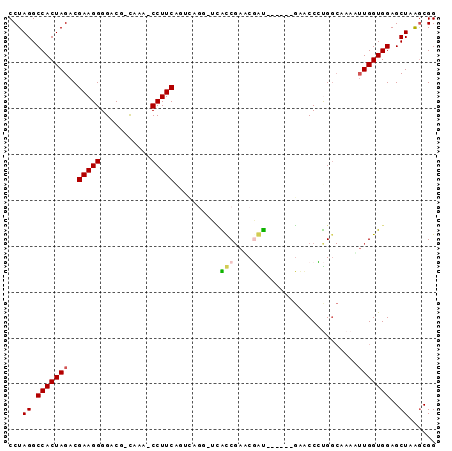
| Location | 3,514,815 – 3,514,901 |
|---|---|
| Length | 86 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -25.10 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514815 86 + 6264404 AGUAAGGCC-AGCGAUGC----UGGCCUUG--GAAUGUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCU ..(((((((-(((...))----))))))))--.........(((((..(((((.......))))).....(((((((...))))))).))))) ( -37.70) >psp.1169 3635914 87 - 5928787 AGGCUCAUCGAACGAU------GAACCCUGGCGAAUUUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU .((.(((((....)))------)).))..(((.......((((((.(((((((.......))))))).)))).))..(((.....)))))).. ( -33.10) >pss.1199 3796958 87 - 6093698 AGGUUCAUCGAACGAU------GAACCCUGGCAAAAUUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU .((((((((....)))------)))))..(((.......((((((.(((((((.......))))))).)))).))..(((.....)))))).. ( -37.30) >pst.1304 4011977 86 - 6397126 AGGAUCACCGAGCGGU------GAACCCUGGCAAA-UUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU .((.(((((....)))------)).))..(((...-...((((((.(((((((.......))))))).)))).))..(((.....)))))).. ( -35.10) >ppk.1135 3939949 89 - 6181863 ----GUGCCGAUCCGCUAACGCGAACCGCGGCAAAAUUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU ----.(((((...(((....))).....)))))........(((((..(((((.......))))).....(((((((...))))))).))))) ( -36.30) >pel.905 4179666 89 - 5888780 ----GUGCCGAUCCGUUUUCACGAACCGCGGCAAAAUUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU ----.(((((....((((....))))..)))))........(((((..(((((.......))))).....(((((((...))))))).))))) ( -31.90) >consensus AGG_UCACCGAACGAU______GAACCCUGGCAAAAUUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCU .....(((((((.......................)))))))((((..(((((.......))))).....(((((((...))))))).)))). (-25.10 = -24.22 + -0.89)

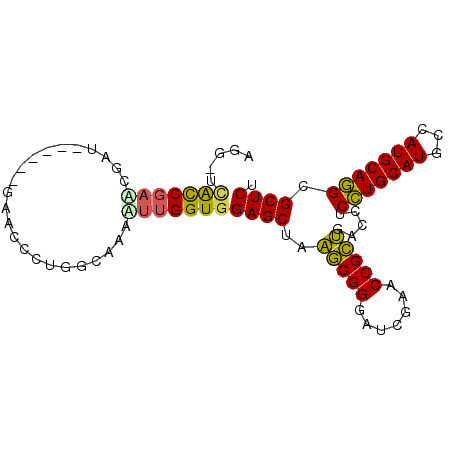
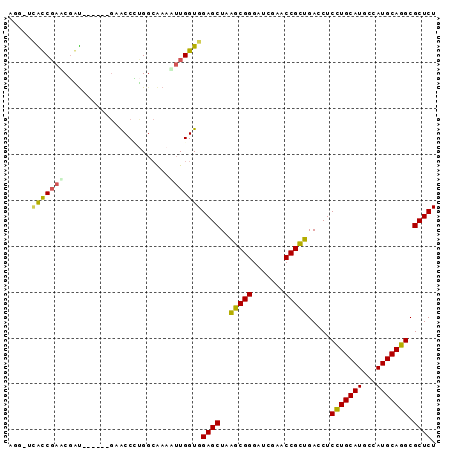
| Location | 3,514,815 – 3,514,901 |
|---|---|
| Length | 86 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -22.80 |
| Energy contribution | -22.67 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514815 86 - 6264404 AGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCACAUUC--CAAGGCCA----GCAUCGCU-GGCCUUACU .((((.(((((((...))))))).....(((((.......)))))..))))..........--.(((((((----((...)))-))))))... ( -35.30) >psp.1169 3635914 87 + 5928787 AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAAUUCGCCAGGGUUC------AUCGUUCGAUGAGCCU .(.(((((.....))).....((((.(.(((((.......))))).).)))).........))).((((((------(((....))))))))) ( -35.30) >pss.1199 3796958 87 + 6093698 AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAUUUUGCCAGGGUUC------AUCGUUCGAUGAACCU .(.(((((.....))).....((((.(.(((((.......))))).).)))).........))).((((((------(((....))))))))) ( -35.00) >pst.1304 4011977 86 + 6397126 AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAA-UUUGCCAGGGUUC------ACCGCUCGGUGAUCCU .(.(((((.....))).....((((.(.(((((.......))))).).)))).....-...)))(((((.(------(((....))))))))) ( -33.00) >ppk.1135 3939949 89 + 6181863 AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAAUUUUGCCGCGGUUCGCGUUAGCGGAUCGGCAC---- .((((.(((((((...))))))).....(((((.......)))))..)))).........(((((..((((((....))))))))))).---- ( -36.30) >pel.905 4179666 89 + 5888780 AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAAUUUUGCCGCGGUUCGUGAAAACGGAUCGGCAC---- .((((.(((((((...))))))).....(((((.......)))))..)))).........(((((..((((((....))))))))))).---- ( -35.80) >consensus AGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAUUUUGCCAGGGUUC______ACCGAUCGGCGA_CCU .(((((((.....))).....((((.(.(((((.......))))).).))))............................))))......... (-22.80 = -22.67 + -0.13)

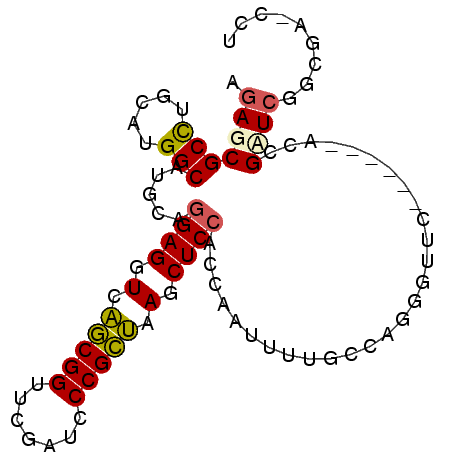
| Location | 3,514,832 – 3,514,941 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -31.39 |
| Energy contribution | -29.36 |
| Covariance contribution | -2.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514832 109 + 6264404 -----UGGCCUUG------GAAUGUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCGAAUUCGCCAUCAGGCGGGC -----((((.(..------(....(((..((((..(((((.......))))).....(((((((...))))))).))))..))).)..)))))..((((.(....)(((....))))))) ( -41.80) >pau.1128 4484193 109 - 6537648 -----UGGCCUUG------GAAUGUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCGAAUUCGCCAUCAGGCGGGC -----((((.(..------(....(((..((((..(((((.......))))).....(((((((...))))))).))))..))).)..)))))..((((.(....)(((....))))))) ( -41.80) >psp.1169 3635930 98 - 5928787 -----GAACCCUG----GCGAAUUUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAAAGCCCCGGAUUUA------------- -----...((..(----((......((..(((((((((((.......)))))))...(((((((...))))))).))))..))(((...)))...)))..)).....------------- ( -37.60) >pf5.1234 4794831 106 - 7074893 UGGCCAAGCCUUGAACUGUAAA-UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCGGAUUUU------------- .(((...))).........(((-((.(.((.(((((((((.......))))).....(((((((...))))))).((((........))))..))))))).))))).------------- ( -32.40) >pfo.1081 4293827 106 - 6438405 UGGCCAGACCUUGAACUGUAAA-UUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAUAGCCCCGGAUUUU------------- (((((((((........))...-((((..((((..(((((.......))))).....(((((((...))))))).))))..))))))).))))..............------------- ( -34.30) >pel.905 4179684 98 - 5888780 -----GAACCGCG----GCAAAAUUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAUAGCCCCGGAUUUC------------- -----...(((.(----((......((..((((..(((((.......))))).....(((((((...))))))).))))..))(((...)))...))).))).....------------- ( -39.10) >consensus _____AAACCUUG____G_AAAUUUGGUGGAGCUAAACGGGAUCGAACCGCCGACCUCCUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCGGAUUUG_____________ .......(((...............)))((.(((((((((.......))))).....(((((((...))))))).((((........))))..)))).)).................... (-31.39 = -29.36 + -2.03)
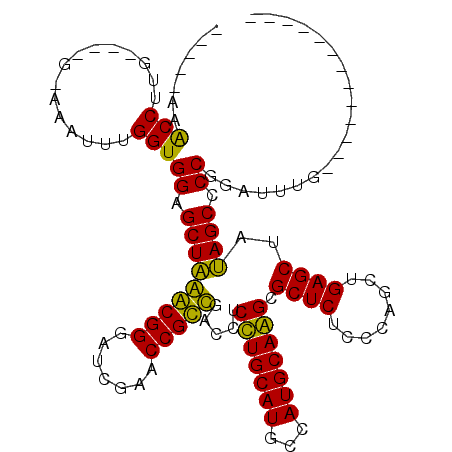
| Location | 3,514,832 – 3,514,941 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -33.26 |
| Energy contribution | -31.53 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514832 109 - 6264404 GCCCGCCUGAUGGCGAAUUCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCACAUUC------CAAGGCCA----- (((((((....)))).....(((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))).........------...)))..----- ( -41.10) >pau.1128 4484193 109 + 6537648 GCCCGCCUGAUGGCGAAUUCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCACAUUC------CAAGGCCA----- (((((((....)))).....(((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))).........------...)))..----- ( -41.10) >psp.1169 3635930 98 + 5928787 -------------UAAAUCCGGGGCUUUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAAUUCGC----CAGGGUUC----- -------------..(((((..(((...(((...)))((..((((.(((((((...))))))).....(((((.......)))))..))))..))......))----).))))).----- ( -37.30) >pf5.1234 4794831 106 + 7074893 -------------AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA-UUUACAGUUCAAGGCUUGGCCA -------------.........(((((.((((.((((((..((((.(((((((...))))))).....(((((.......)))))..))))..))..-.....))))...))))))))). ( -35.11) >pfo.1081 4293827 106 + 6438405 -------------AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAA-UUUACAGUUCAAGGUCUGGCCA -------------.....(((((.((..((((....(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).-.....))))..)).)))))... ( -35.80) >pel.905 4179684 98 + 5888780 -------------GAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAAUUUUGC----CGCGGUUC----- -------------.....(((.(((...(((...)))((..((((.(((((((...))))))).....(((((.......)))))..))))..))......))----).)))...----- ( -37.90) >consensus _____________AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAAGAGGUCAACGGUUCGAUCCCGCCUAGCUCCACCAAAUUC_C____CAAGGCCA_____ ..................((.((((....))))...(((..((((.(((((((...))))))).....(((((.......)))))..))))..)))..............))........ (-33.26 = -31.53 + -1.72)
| Location | 3,514,861 – 3,514,981 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -24.98 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514861 120 - 6264404 GUGUCGACGACCGGAAGGUUGUCGAAGCGAUCGGAAGAUCGCCCGCCUGAUGGCGAAUUCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUC ...(((((((((....))))))))).((((((....)))))).((((....))))...(((((((....))))(((((...((.(.(((((((...))))))).).))..)))))))).. ( -59.00) >pf5.1234 4794870 88 + 7074893 AUGUCGCUGCGCCG--------CGAGGCG------------------------AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUC ....((((.((((.--------...))))------------------------....((((((((....)))...)))))..))))(((((((...))))))).((((((....)))))) ( -32.40) >pfo.1081 4293866 89 + 6438405 AUGUCGCUCCGCCG--------AGAGGCGA-----------------------AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUC ...(((((.((((.--------...)))).-----------------------....((((((((....)))...))))).((.(.(((((((...))))))).).)))))))....... ( -33.80) >pst.1304 4012023 89 + 6397126 GUUGAUCUCCGCCG--------CGAGGCGA-----------------------UAAAUCCGGGGCUUUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUC (((((((((((((.--------...)))..-----------------------.......((.((((...(((....))).)))).))(((((...)))))))))))))))......... ( -38.40) >ppk.1135 3939998 89 + 6181863 AUGUCGCUGGGCCG--------CGAGGCUA-----------------------GAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUC ...((((((((((.--------...)))).-----------------------.....((.((((....)))).(((.....))).(((((((...))))))).)).))))))....... ( -34.50) >pel.905 4179715 89 + 5888780 AUGUCGCUGGGCCG--------AGAGGCUA-----------------------GAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUC ...((((((((((.--------...)))).-----------------------.....((.((((....)))).(((.....))).(((((((...))))))).)).))))))....... ( -34.00) >consensus AUGUCGCUGCGCCG________CGAGGCGA_______________________AAAAUCCGGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUC ..((((...((((............)))).............................(((.((((...((((........)))).(((((((...))))))).))))..)))..)))). (-24.98 = -25.23 + 0.25)
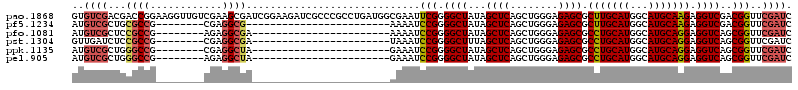
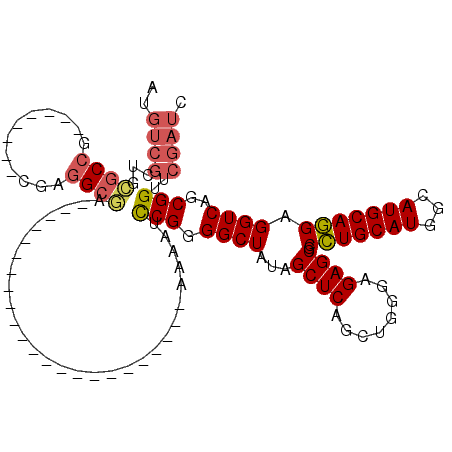
| Location | 3,514,901 – 3,515,021 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.69 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -16.67 |
| Energy contribution | -17.87 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514901 120 - 6264404 GACGCCACUUCAUGUUCAUCCGUCCCAGCAAUGGAUGAACGUGUCGACGACCGGAAGGUUGUCGAAGCGAUCGGAAGAUCGCCCGCCUGAUGGCGAAUUCGGGGCUAUAGCUCAGCUGGG ....(((((.(((((((((((((.......)))))))))))))(((((((((....))))))))).((((((....)))))).((((....))))......((((....)))))).))). ( -60.30) >pf5.1234 4794910 79 + 7074893 GACGCCAGU---------UUCACACAAGCGAUGUUGCAAGAUGUCGCUGCGCCG--------CGAGGCG------------------------AAAAUCCGGGGCUAUAGCUCAGCUGGG ....(((((---------(.......(((((((((....))))))))).((((.--------...))))------------------------........((((....)))))))))). ( -31.10) >pfo.1081 4293906 80 + 6438405 GACGCCAGU---------UUUACAGAAGUGAUGUUGCAAGAUGUCGCUCCGCCG--------AGAGGCGA-----------------------AAAAUCCGGGGCUAUAGCUCAGCUGGG ....(((((---------(...(.(((((((((((....))))))))).((((.--------...)))).-----------------------....)).)((((....)))))))))). ( -28.40) >pst.1304 4012063 79 + 6397126 GACGCCAGU---------UUCAC-CGAGUGAUACUUCAACGUUGAUCUCCGCCG--------CGAGGCGA-----------------------UAAAUCCGGGGCUUUAGCUCAGCUGGG ....(((((---------((((.-((..(((....))).)).)))....((((.--------...)))).-----------------------........((((....)))))))))). ( -25.10) >ppk.1135 3940038 78 + 6181863 GACGCCAGU---------UUUACACAAGCGAUGUUUUA--AUGUCGCUGGGCCG--------CGAGGCUA-----------------------GAAAUCCGGGGCUAUAGCUCAGCUGGG ....(((((---------(.......((((((((....--)))))))).((((.--------...)))).-----------------------........((((....)))))))))). ( -28.30) >pel.905 4179755 79 + 5888780 GACGCCAGU---------UUUGCACAAGCGAUGUUUAG-CAUGUCGCUGGGCCG--------AGAGGCUA-----------------------GAAAUCCGGGGCUAUAGCUCAGCUGGG ....(((((---------(.......((((((((....-.)))))))).((((.--------...)))).-----------------------........((((....)))))))))). ( -27.60) >consensus GACGCCAGU_________UUCACACAAGCGAUGUUUCAACAUGUCGCUGCGCCG________CGAGGCGA_______________________AAAAUCCGGGGCUAUAGCUCAGCUGGG ....(((((.................((((((((......)))))))).((((............))))................................((((....)))).))))). (-16.67 = -17.87 + 1.20)
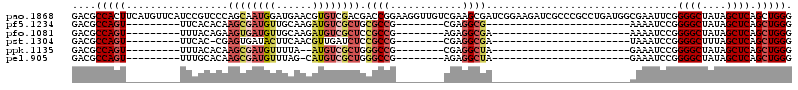
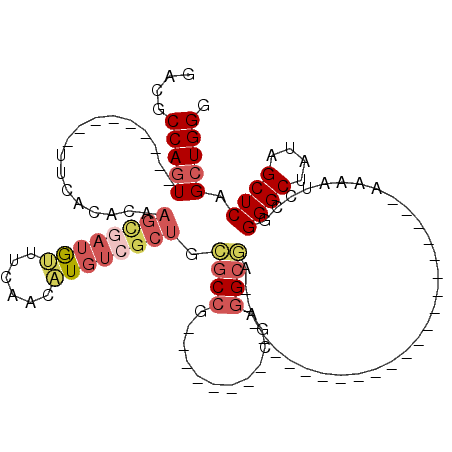
| Location | 3,514,941 – 3,515,061 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.18 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -24.15 |
| Energy contribution | -25.32 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514941 120 - 6264404 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUCAUGUUCAUCCGUCCCAGCAAUGGAUGAACGUGUCGACGACCGGAAGGUUGUCGAAGCGAUCGGAAGAUC ((((((..((....))...))))))....(((((....)))))((.....(((((((((((((.......)))))))))))))(((((((((....))))))))).))((((....)))) ( -58.70) >pss.1199 3797072 91 + 6093698 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUCAC-CGAGUGAUGCCGGAAGGUUGAUCUCCGCCG--------AGAGGCG----------- (((...((.(((((......((((..(..(((((....)))))....).---------.)).)-)(((....(((....)))....))))))))--------.))))).----------- ( -38.50) >pf5.1234 4794937 92 + 7074893 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUCACACAAGCGAUGUUGCAAGAUGUCGCUGCGCCG--------CGAGGCG----------- (((...((.(((((......((..((.(.(((((....))))).).)).---------.)).....(((((((((....))))))))).)))))--------.))))).----------- ( -38.30) >pfo.1081 4293933 93 + 6438405 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUACAGAAGUGAUGUUGCAAGAUGUCGCUCCGCCG--------AGAGGCGA---------- (((...((.(((((((((....(((....(((((....))))))))...---------.))))...(((((((((....))))))))).)))))--------.)))))..---------- ( -36.90) >ppk.1135 3940065 91 + 6181863 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUACACAAGCGAUGUUUUA--AUGUCGCUGGGCCG--------CGAGGCUA---------- (((...((.((((.((((....(((....(((((....))))))))...---------.)))).(.((((((((....--)))))))).)))))--------.)))))..---------- ( -35.80) >pel.905 4179782 92 + 5888780 GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUGCACAAGCGAUGUUUAG-CAUGUCGCUGGGCCG--------AGAGGCUA---------- (((.((((..((((....((((((.......))))).)....))))...---------...((...((((((((....-.))))))))..)).)--------))))))..---------- ( -35.50) >consensus GCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU_________UUCACACAAGCGAUGUUGCAAGAUGUCGCUGCGCCG________AGAGGCGA__________ (((.......((((....((((((.......))))).)....))))....................((((((((......)))))))).................)))............ (-24.15 = -25.32 + 1.17)
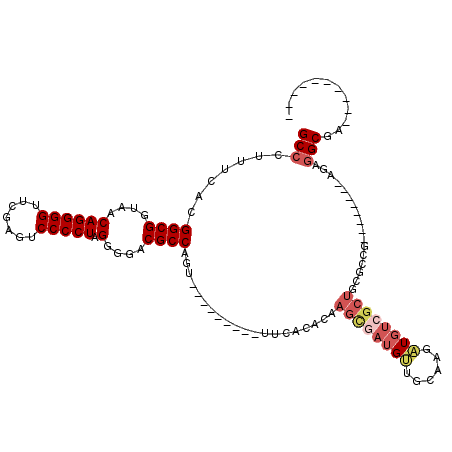
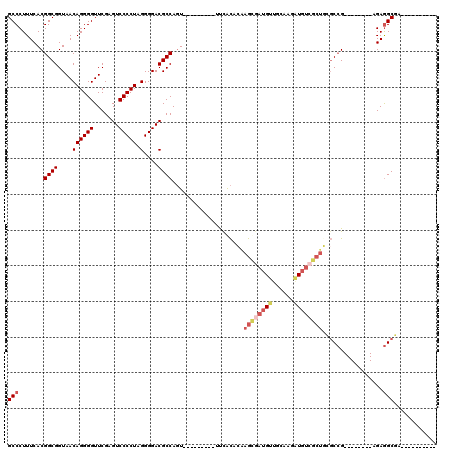
| Location | 3,514,981 – 3,515,100 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.27 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -38.37 |
| Energy contribution | -38.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514981 119 + 6264404 GUUCAUCCAUUGCUGGGACGGAUGAACAUGAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-AAACCUUCG ((((((((...........))))))))..((((.(.(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))-...))))). ( -54.70) >pau.1128 4484342 119 - 6537648 GCUCAUCCAUUGCUGGGACGGAUGAACAUGAAGUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-AAACCUUCG (.((((((...........)))))).)..((((.(.(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))-...))))). ( -50.50) >pf5.1234 4794958 111 - 7074893 CUUGCAACAUCGCUUGUGUGAA---------ACUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCAAACCCUUC .........((((....)))).---------....((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))))......... ( -46.00) >pfo.1081 4293955 111 - 6438405 CUUGCAACAUCACUUCUGUAAA---------ACUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCAAACCCUUC .(((((..........))))).---------....((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))))......... ( -43.60) >ppk.1135 3940087 107 - 6181863 --UAAAACAUCGCUUGUGUAAA---------ACUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG--AACCUUCU --.......(((((.((.....---------)).)))((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))--)....... ( -42.00) >pel.905 4179804 108 - 5888780 G-CUAAACAUCGCUUGUGCAAA---------ACUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG--AACCUUCU .-.......(((((.((.....---------)).)))((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))--)....... ( -42.00) >consensus GUUCAAACAUCGCUUGUGCAAA_________ACUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG_AAACCUUCC ..................................(((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))....)).... (-38.37 = -38.37 + 0.00)

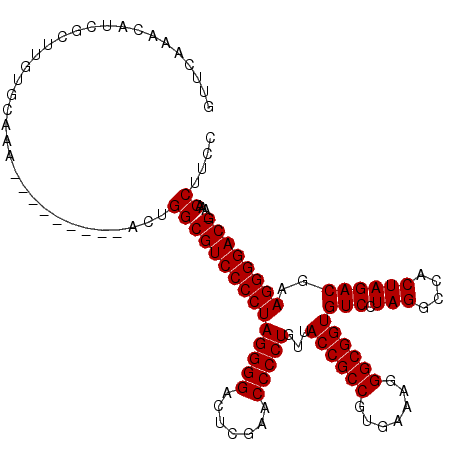
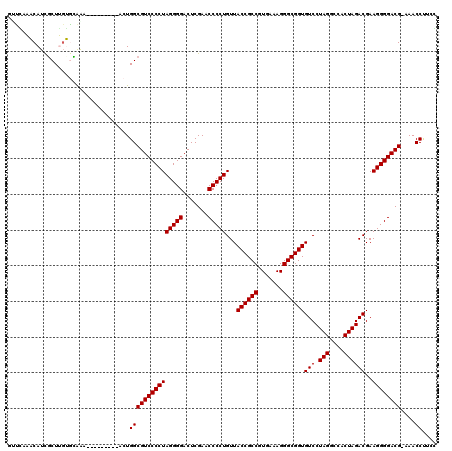
| Location | 3,514,981 – 3,515,100 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.27 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -37.23 |
| Energy contribution | -37.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3514981 119 - 6264404 CGAAGGUUU-CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUCAUGUUCAUCCGUCCCAGCAAUGGAUGAAC .((((....-((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))...))))..((((((((((.......)))))))))) ( -53.00) >pau.1128 4484342 119 + 6537648 CGAAGGUUU-CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCACUUCAUGUUCAUCCGUCCCAGCAAUGGAUGAGC .((((....-((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))...))))..((((((((((.......)))))))))) ( -53.00) >pf5.1234 4794958 111 + 7074893 GAAGGGUUUGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUCACACAAGCGAUGUUGCAAG ((((.....(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))...)---------)))......(((....)))... ( -42.10) >pfo.1081 4293955 111 + 6438405 GAAGGGUUUGCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUACAGAAGUGAUGUUGCAAG .........(((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))(((.---------.((((....))))..))).... ( -42.60) >ppk.1135 3940087 107 + 6181863 AGAAGGUU--CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUACACAAGCGAUGUUUUA-- ....((((--((.(((((..((((((....))).)))((((((.......))))))...)))))..)))(((((....)))))))).((---------((.....)))).........-- ( -39.00) >pel.905 4179804 108 + 5888780 AGAAGGUU--CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU---------UUUGCACAAGCGAUGUUUAG-C ....((((--((.(((((..((((((....))).)))((((((.......))))))...)))))..)))(((((....))))))))...---------.((((....)))).......-. ( -40.90) >consensus AGAAGGUUU_CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAGU_________UUCACACAAGCGAUGUUUCAAC ....((....((((((((..((((((....))).)))((((((.......))))))...(((((.......))))))))))))))).................................. (-37.23 = -37.23 + -0.00)

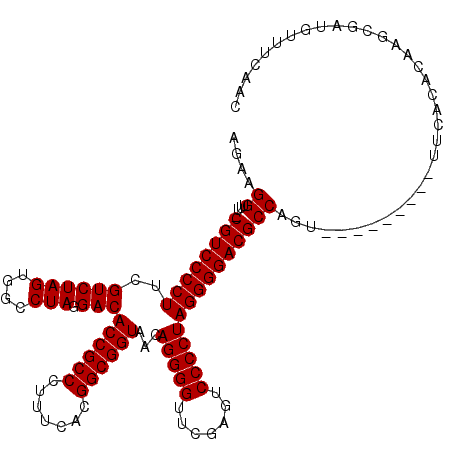
| Location | 3,515,021 – 3,515,124 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -38.75 |
| Consensus MFE | -28.60 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3515021 103 + 6264404 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG-AAACCUUCGUAGGAAUCCGCAAAACCAA-------------CCG--CG .((((((((.......)))))..(((((((.......)))))))..((((....))))(((((((.....-...))))))))))....(((........-------------..)--)) ( -36.50) >psp.1169 3636080 107 - 5928787 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACA-AAACCUUCUUA--------CAAAACAAACGAUCGCUGGAU-CCG--CU ...((((((.......))))))..((((((.......))))))(((.(((....))))))(((((.....-...)))))...--------..........((((....)))-)..--.. ( -33.90) >pf5.1234 4794989 109 - 7074893 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCAAACCCUUCUG--------UACAACUGACCGGUGCUGAGUACCG--AU ...((((((.......))))))..((((((.......))))))(((((((....))))..((((((........))))))..--------.......)))((((((...))))))--.. ( -41.80) >pfo.1081 4293986 109 - 6438405 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGCAAACCCUUCUA--------UACAACUGAUCAACGCUGAGUGUUG--AU ...((((((.......))))))..((((((.......))))))(((.(((....))))))((((((........))))))..--------........((((((((...))))))--)) ( -41.00) >ppk.1135 3940116 110 - 6181863 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG--AACCUUCUUA------CCUUCAAGACCCGGUUGCUGGAC-CCGGUCU ...((((((.......))))))..((((((.......))))))(((.(((....))))))(((((..(.(--(....)))..------))))).(((((.((((....)))-).))))) ( -41.80) >pel.905 4179834 110 - 5888780 CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG--AACCUUCUUA------CCUUCAAGACCCGCUUACUGGAA-CCGGCCU ...((((((.......)))))).(((((((.......)))))))....(((((.......(((((.....--..)))))...------((...(((.....)))...))..-..))))) ( -37.50) >consensus CCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG_AAACCUUCUUA________UACAACAAACGAUUGCUGGAU_CCG__CU (((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))..................................................... (-28.60 = -28.60 + -0.00)

| Location | 3,515,124 – 3,515,208 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -29.57 |
| Energy contribution | -28.07 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3515124 84 + 6264404 UAUUCCC----------------------------------GGCAAU--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAU ......(----------------------------------(((...--.(((..((((..(((((.......))))).....(((((((...))))))).))))..)))))))...... ( -30.70) >pf5.1234 4795098 109 - 7074893 CGAUCCA---------AGGCCGGUGUGGCCAGGCCUUGAACUGUAAA--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAU .....((---------(((((((.....)).))))))).........--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))......... ( -42.40) >pfo.1081 4294095 109 - 6438405 CGCUUCG---------AAGCCGGUGUGGCCAGGCUUCGAAGUGUAAA--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAU (((((((---------(((((((.....)).))))))))))))....--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))......... ( -50.30) >pst.1304 4012250 120 - 6397126 UGUUUCACAUCGAAUCAGAACAAUGUAGCUGCGCUCUGAAACGCUAAAUUUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAA .((((((...((...(((..........)))))...))))))(((....((((..(((((((((((.......)))))))...(((((((...))))))).))))..))))...)))... ( -40.60) >ppk.1135 3940226 101 - 6181863 UGCUUAUCAGCCUUA-----------------GCUGCUAAGCGGAAU--UUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAU .(((((.((((....-----------------)))).))))).....--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))......... ( -40.50) >pel.905 4179944 101 - 5888780 UGUCUGACAGCUUUU-----------------GCUACCAGUCGGAAU--UUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAU ..(((((((((....-----------------)))....))))))..--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))......... ( -38.00) >consensus UGUUUCA_________________________GCUUCGAAGCGCAAA__UUGGUGGAGCUAAACGGGAUCGAACCGCCGACCUCCUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAU .................................................((((..((((..(((((.......))))).....(((((((...))))))).))))..))))......... (-29.57 = -28.07 + -1.50)
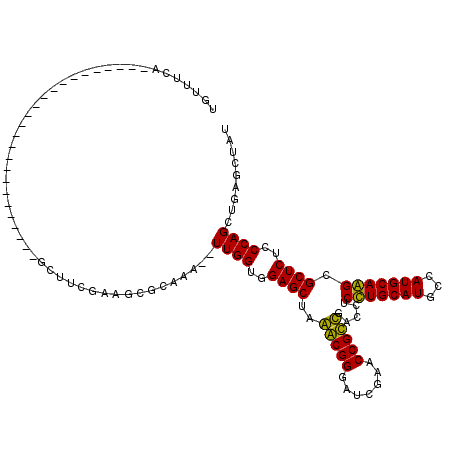
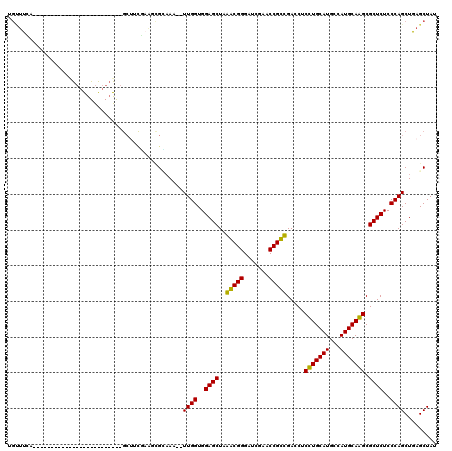
| Location | 3,515,124 – 3,515,208 |
|---|---|
| Length | 84 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.38 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -27.85 |
| Energy contribution | -26.35 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3515124 84 - 6264404 AUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--AUUGCC----------------------------------GGGAAUA ....(((.(((((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--...)).----------------------------------))).... ( -28.80) >pf5.1234 4795098 109 + 7074893 AUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--UUUACAGUUCAAGGCCUGGCCACACCGGCCU---------UGGAUCG ..........(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--......((((((((((.((.....)))))))---------))))).. ( -42.00) >pfo.1081 4294095 109 + 6438405 AUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--UUUACACUUCGAAGCCUGGCCACACCGGCUU---------CGAAGCG ..........(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--......((((((((((.((.....)))))))---------))))).. ( -43.60) >pst.1304 4012250 120 + 6397126 UUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAAUUUAGCGUUUCAGAGCGCAGCUACAUUGUUCUGAUUCGAUGUGAAACA ..((((....(((..((((.(((((((...))))))).....(((((.......)))))..))))..)))......(((((....)))))))))((((((........))))))...... ( -41.10) >ppk.1135 3940226 101 + 6181863 AUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAA--AUUCCGCUUAGCAGC-----------------UAAGGCUGAUAAGCA ..........(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--.....(((((.((((-----------------....)))).))))). ( -39.50) >pel.905 4179944 101 + 5888780 AUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAA--AUUCCGACUGGUAGC-----------------AAAAGCUGUCAGACA ..........(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--.......(((..(((-----------------....)))..)))... ( -36.50) >consensus AUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAAGAGGUCAACGGUUCGAUCCCGCCUAGCUCCACCAA__AUUACGCUUCGAAGC_________________________UGAAACA ..........(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).................................................. (-27.85 = -26.35 + -1.50)
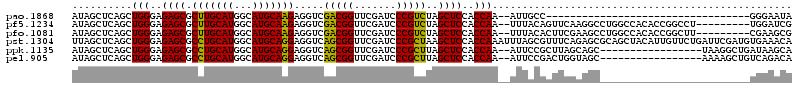
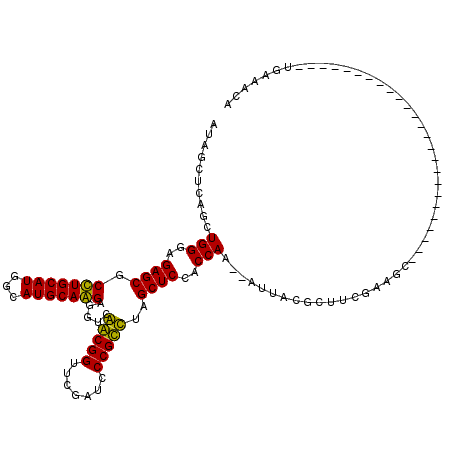
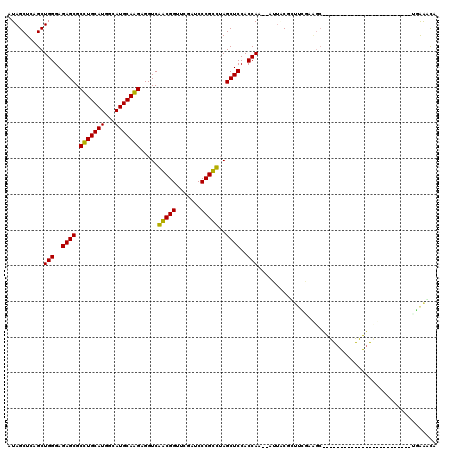
| Location | 3,515,131 – 3,515,218 |
|---|---|
| Length | 87 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -37.43 |
| Consensus MFE | -30.83 |
| Energy contribution | -29.50 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3515131 87 + 6264404 -------------------------------GGCAAU--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCAAAG -------------------------------(((...--..((..((((..(((((.......))))).....(((((((...))))))).))))..))(((...)))...)))...... ( -31.20) >pf5.1234 4795105 111 - 7074893 ------AGGCCGGUGUGGCCAGGCCUUGAACUGUAAA--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCCGGA- ------...(((((((((((((((........))...--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))))).))))))...)))).- ( -41.80) >pfo.1081 4294102 108 - 6438405 ------AAGCCGGUGUGGCCAGGCUUCGAAGUGUAAA--UUGGUGGAGCUAGACGGGAUCGAACCGUCGACCUCUUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCC---- ------.....((((((((((((((....))).....--((((..((((..(((((.......))))).....(((((((...))))))).))))..))))))).)))))).))..---- ( -39.60) >pst.1304 4012260 119 - 6397126 CGAAUCAGAACAAUGUAGCUGCGCUCUGAAACGCUAAAUUUGGUGGAGCUUAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAAAGCCCCACA- ...............((((((((........))).....((((..(((((((((((.......)))))))...(((((((...))))))).))))..))))...)))))..........- ( -39.00) >ppk.1135 3940236 97 - 6181863 CCUUA-----------------GCUGCUAAGCGGAAU--UUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAUAGCCCC---- .....-----------------...(((((((...(.--((((..((((..(((((.......))))).....(((((((...))))))).))))..)))).)..))).))))...---- ( -36.70) >pel.905 4179954 97 - 5888780 CUUUU-----------------GCUACCAGUCGGAAU--UUGGUGGAGCUAAGCGGGAUCGAACCGCUGACCUCCUGCAUGCCAUGCAGGCGCUCUCCCAGCUGAGCUAUAGCCCC---- .....-----------------((((..((((((...--.(((..((((..(((((.......))))).....(((((((...))))))).))))..))).)))).)).))))...---- ( -36.30) >consensus ______________________GCUUCGAAGCGCAAA__UUGGUGGAGCUAAACGGGAUCGAACCGCCGACCUCCUGCAUGCCAUGCAAGCGCUCUCCCAGCUGAGCUAUAGCCCC____ ........................................(((..((((..(((((.......))))).....(((((((...))))))).))))..)))((((.....))))....... (-30.83 = -29.50 + -1.33)
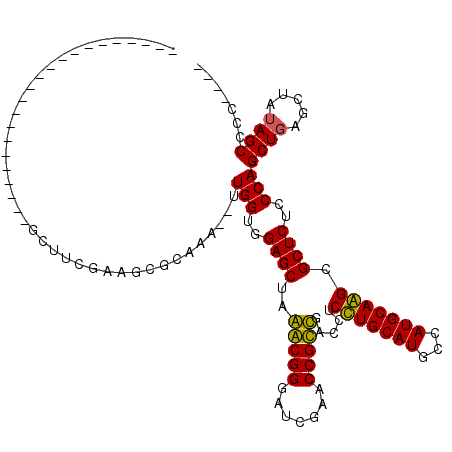
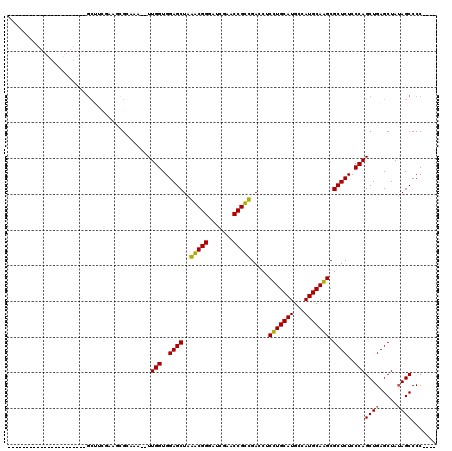
| Location | 3,515,131 – 3,515,218 |
|---|---|
| Length | 87 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -32.68 |
| Energy contribution | -30.90 |
| Covariance contribution | -1.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1868 3515131 87 - 6264404 CUUUGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--AUUGCC------------------------------- ...((((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))))....--......------------------------------- ( -30.60) >pf5.1234 4795105 111 + 7074893 -UCCGGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--UUUACAGUUCAAGGCCUGGCCACACCGGCCU------ -....(((((..((((....(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--.....))))...)))))((((.....)))).------ ( -40.20) >pfo.1081 4294102 108 + 6438405 ----GGGGCUAUAGCUCAGCUGGGAGAGCGCUUGCAUGGCAUGCAAGAGGUCGACGGUUCGAUCCCGUCUAGCUCCACCAA--UUUACACUUCGAAGCCUGGCCACACCGGCUU------ ----.((((....))))...(((..((((.(((((((...))))))).....(((((.......)))))..))))..))).--...........(((((.((.....)))))))------ ( -37.20) >pst.1304 4012260 119 + 6397126 -UGUGGGGCUUUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUAAGCUCCACCAAAUUUAGCGUUUCAGAGCGCAGCUACAUUGUUCUGAUUCG -.(((((((((..((((........)))).(((((((...))))))).....(((((.......)))))))))))))).........((..((((((((.........))))))))..)) ( -44.80) >ppk.1135 3940236 97 + 6181863 ----GGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAA--AUUCCGCUUAGCAGC-----------------UAAGG ----.((((....))))(((((...(((((((.....))).....((((.(.(((((.......))))).).)))).....--.....))))..))))-----------------).... ( -36.60) >pel.905 4179954 97 + 5888780 ----GGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAGGAGGUCAGCGGUUCGAUCCCGCUUAGCUCCACCAA--AUUCCGACUGGUAGC-----------------AAAAG ----(((((((..((((........)))).(((((((...))))))).....(((((.......))))))))))))((((.--........))))...-----------------..... ( -34.00) >consensus ____GGGGCUAUAGCUCAGCUGGGAGAGCGCCUGCAUGGCAUGCAAGAGGUCAACGGUUCGAUCCCGCCUAGCUCCACCAA__AUUACGCUUCGAAGC______________________ ....(((((((..((((........)))).(((((((...))))))).....(((((.......))))))))))))............................................ (-32.68 = -30.90 + -1.78)
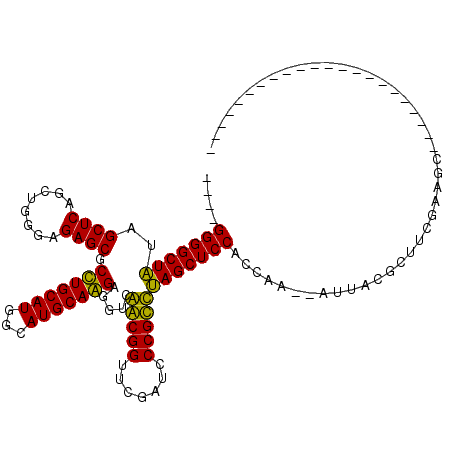
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:33 2007