
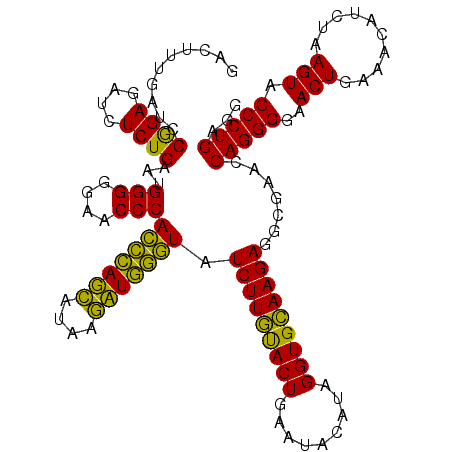
| Sequence ID | pao.3201 |
|---|---|
| Location | 6,042,522 – 6,042,984 |
| Length | 462 |
| Max. P | 0.999993 |

| Location | 6,042,522 – 6,042,642 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -41.32 |
| Energy contribution | -38.60 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.37 |
| Structure conservation index | 1.06 |
| SVM decision value | 5.77 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
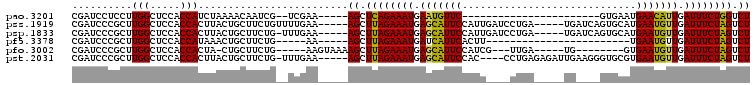
>pao.3201 6042522 120 - 6264404 GACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..........((((....))))...(((....)))(((((((....))))))).(((((((((.........))))))))).......(((((..(((..........))).)))))... ( -39.70) >pau.3340 6315441 120 - 6537648 GACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..........((((....))))...(((....)))(((((((....))))))).(((((((((.........))))))))).......(((((..(((..........))).)))))... ( -39.70) >pel.69 5771056 120 + 5888780 GACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..........((((....))))...(((....)))(((((((....))))))).(((((((((.........))))))))).......(((((..(((..........))).)))))... ( -40.80) >pf5.3378 6385680 120 - 7074893 GACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..........((((....))))...(((....)))(((((((....))))))).(((((((((.........))))))))).......(((((..(((..........))).)))))... ( -37.80) >pfo.3002 5729300 120 - 6438405 GACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..(((((...))))).((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))...(((((..(((..........))).)))))... ( -38.50) >pst.2031 3872392 120 - 6397126 GACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG .........((.....((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))...(((((..(((..........))).))))).)) ( -38.00) >consensus GACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAAGAUGGGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGG ..........((((....))))...(((....)))(((((((....))))))).(((((((((.........))))))))).......(((((..(((..........))).)))))... (-41.32 = -38.60 + -2.72)


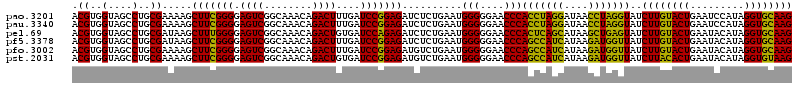
| Location | 6,042,562 – 6,042,682 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -40.68 |
| Consensus MFE | -42.97 |
| Energy contribution | -40.12 |
| Covariance contribution | -2.86 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.13 |
| Structure conservation index | 1.06 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042562 120 - 6264404 ACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAG .((..(....)..))......((((((((.((((((((.....))))..))))...)))))))).(((....)))(((((((....)))))))..((((((((.........)))))))) ( -42.10) >pau.3340 6315481 120 - 6537648 ACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAG .((..(....)..))......((((((((.((((((((.....))))..))))...)))))))).(((....)))(((((((....)))))))..((((((((.........)))))))) ( -42.10) >pel.69 5771096 120 + 5888780 ACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAG .((..(....)..)).....(((((((.((((........))))....)))))))..........(((....)))(((((((....)))))))..((((((((.........)))))))) ( -40.40) >pf5.3378 6385720 120 - 7074893 ACGUGGUAGCCUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAG .((..(....)..))......((((((((.((((((((.....))))..))))...)))))))).(((....)))(((((((....)))))))..((((((((.........)))))))) ( -40.20) >pfo.3002 5729340 120 - 6438405 ACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAG .((..(....)..)).....((((((((((((........)))))...)))))))..........(((....)))(((((((....)))))))..((((((((.........)))))))) ( -40.00) >pst.2031 3872432 120 - 6397126 ACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAG .((..(....)..)).....(((((((.((((........))))....)))))))..........(((....)))(((((((....)))))))..((((((((.........)))))))) ( -39.30) >consensus ACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCCAGCAUAAGAUGGGUAUCUUGUACUGAAUACAUAGGUGCAAG .((..(....)..)).....(((((((.((((........))))....)))))))..........(((....)))(((((((....)))))))..((((((((.........)))))))) (-42.97 = -40.12 + -2.86)
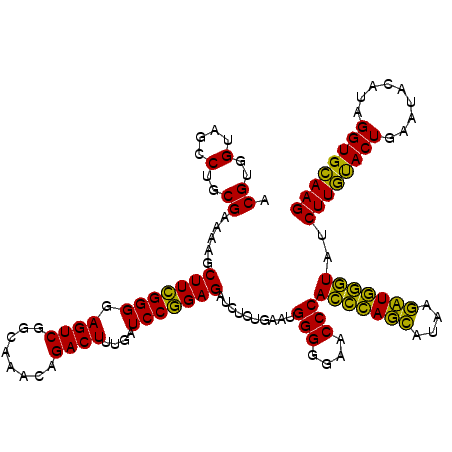
| Location | 6,042,682 – 6,042,794 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.54 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -23.82 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042682 112 + 6264404 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCG-------- .((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..)))).......-------- ( -24.50) >pau.3340 6315601 112 + 6537648 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCG-------- .((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..)))).......-------- ( -24.50) >pel.69 5771216 120 - 5888780 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCAUUUCGCUC .((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..))))....((.....)).. ( -25.30) >pf5.3378 6385840 117 + 7074893 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUAUGAAGACGACAUUCGCCGAAAAUUUGCA-UUC--UU .((((...((((.((.....))))))......((.(((((((((......((((((.((...........)).))))))......)))).)).))).))..)))).......-...--.. ( -21.30) >pfo.3002 5729460 118 + 6438405 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGAAUUUC--UC .((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..))))...........--.. ( -24.50) >pst.2031 3872552 112 + 6397126 CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAGUUUGCA-------- (((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..)))))......-------- ( -26.80) >consensus CUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCCGAAAAUUCGCA________ .((((...((((.((.....))))))......((.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).))..))))............... (-23.82 = -23.98 + 0.17)

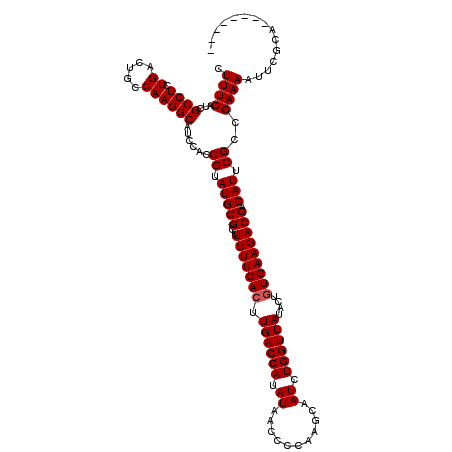
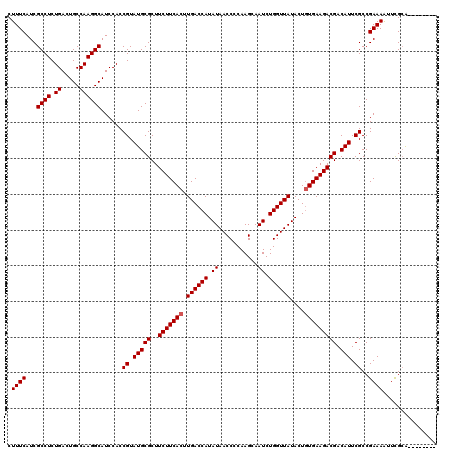
| Location | 6,042,762 – 6,042,861 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 74.14 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -18.37 |
| Energy contribution | -16.99 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042762 99 - 6264404 GAAG---UAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUCA------AG----------CGCGAAUUUUCGGCGAAUGUCGUCUUCACAGU ((((---...((((((((((((.((......)).)))))))).)))).....((((((((((......------..----------))))))))))((((....)))).))))..... ( -31.20) >pau.3340 6315681 99 - 6537648 GAAG---UAAGACUGAAUGAUCUCUUUCACUGGUGAUCAUUCAAGUCAAGGUAAAAUUUGCGAGUUCA------AG----------CGCGAAUUUUCGGCGAAUGUCGUCUUCACAGU ((((---...((((((((((((.((......)).)))))))).)))).....((((((((((......------..----------))))))))))((((....)))).))))..... ( -31.20) >pel.69 5771296 113 + 5888780 GAA----AUAGACUGAAUG-CCAGUUUCACUGCUGGUUAUUCAGGCUAAGGUAAAAUUUGUGAGUUCUGCUCUUAAUUGAGCGAAAUGCGAAUUUUCGGCGAAUGUCGUCUUCACAGU (((----.(((.(((((((-(((((......))))).))))))).)))..(.(((((((((...(((.((((......)))))))..))))))))))((((.....)))))))..... ( -36.10) >psp.1833 3634635 95 - 5928787 GAAAGAAAUAGACCGGGCA-CCUCUUUCACUGGUGCGUGUCCGGGCUAAGGUAAAGUUUGUG------------AA----------UGCAAACUUUCGGCGAAUGUCGUCUUCACAGU ....(((.(((.(((((((-((.((......)).).)))))))).)))..(.(((((((((.------------..----------.))))))))))((((.....)))))))..... ( -31.10) >pf5.3378 6385920 111 - 7074893 GAAAG--AUAGACUGAACG-UUACUUUCACUGGUAACGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCU-CUUUUAAUUAA--GAA-UGCAAAUUUUCGGCGAAUGUCGUCUUCAUAGU (((..--.(((.((((.((-(((((......)))))))..)))).)))..(.(((((((((..(((((-..........)--)))-)))))))))))((((.....)))))))..... ( -26.90) >pfo.3002 5729540 110 - 6438405 GAAAG--AUAGACUGAACG-UUACUUUCACUGGUAACGGAUCAGGCUAAGGUAAAAUUUGUGAGUUCU-CUU--AAUUGA--GAAAUUCGAAUUUUCGGCGAAUGUCGUCUUCACAGU (((..--.(((.((((.((-(((((......)))))))..)))).)))..(.((((((((....((((-(..--....))--)))...)))))))))((((.....)))))))..... ( -30.20) >consensus GAAA___AUAGACUGAACG_UCACUUUCACUGGUGAUCAUUCAGGCUAAGGUAAAAUUUGUGAGUUCU______AA__________UGCGAAUUUUCGGCGAAUGUCGUCUUCACAGU (((.....(((.(((((((.((.((......)).)).))))))).)))..(.((((((((((........................)))))))))))((((.....)))))))..... (-18.37 = -16.99 + -1.38)

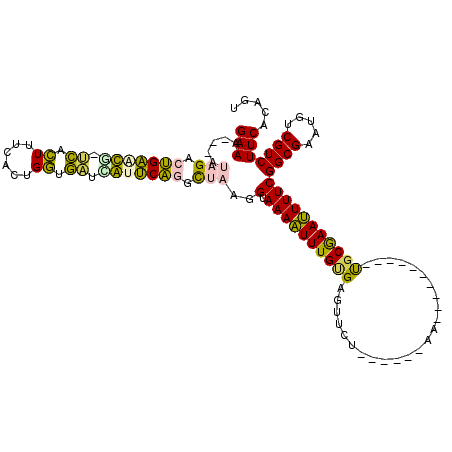
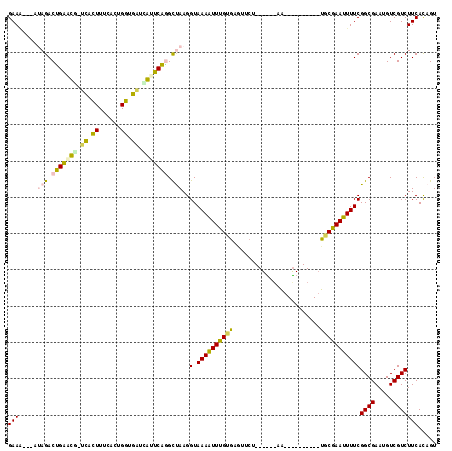
| Location | 6,042,861 – 6,042,950 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -17.27 |
| Consensus MFE | -8.71 |
| Energy contribution | -7.88 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042861 89 + 6264404 UAUCACAUACCCGAAUUUUUAAAGAACAGUUCUGGUGCAAAGACCAGAAAUCAAUGUUCAUUCAC-----------------------GAACAUUCAUUUCUGAGCU-----UUCGA .......((((.(((((.((....)).))))).)))).((((..(((((((.(((((((......-----------------------))))))).)))))))..))-----))... ( -23.40) >pss.1919 3850729 105 + 6093698 UAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAUC-AAAGACUAGAAAUCAACAUUCAUGCACUGAUCA-----UCAGGAUCAAUGGAAUGCUCAUUUCUAAGCU-----UUCAA ...........................-.........-((((..(((((((...(((((...((.(((((.-----....))))).)))))))...)))))))..))-----))... ( -16.70) >psp.1833 3634730 105 + 5928787 UAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAUC-AAAGACUAGAAAUCAACAUUCAUGCACUGAUCA-----UCAGGAUCAAUGGAAUGCUCAUUUCUAAGCU-----UUCAA ...........................-.........-((((..(((((((...(((((...((.(((((.-----....))))).)))))))...)))))))..))-----))... ( -16.70) >pf5.3378 6386031 84 + 7074893 UAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAUCAAAAGACUAGAAAUCAACAUUCA------------------------AAGUGAAUGAUCAUUUCUAAGCU-----UU--- ...........................-..........((((..(((((((...(((((.------------------------....)))))...)))))))..))-----))--- ( -11.80) >pfo.3002 5729650 97 + 6438405 UAUCACAUACCCAAAUUUUUAAAGAAC-GAACCAGUCAAAAGACUAGAAAUCAACAUUCAC--------CA-----UCAA---CGAUGGAAUGCUCAUUUCUAAGCUUUUACUU--- ...........................-.....(((.(((((..(((((((...((((..(--------((-----((..---.)))))))))...)))))))..)))))))).--- ( -15.90) >pst.2031 3872730 106 + 6397126 UAUCACAUACCCAAAUUUUUAAAGAAC-GAUCUAAUC-AAAGACUAGAAAUCAACAUUCACGCACCCUUCAAUCUCUCAGG----GUGGAAUGCUCAUUUCUAAGCU-----UUCAA ...........................-.........-((((..(((((((...(((((...((((((..........)))----))))))))...)))))))..))-----))... ( -19.10) >consensus UAUCACAUACCCAAAUUUUUAAAGAAC_GAUCUAAUC_AAAGACUAGAAAUCAACAUUCAUGCAC____CA_____UCAG____AAUGGAAUGCUCAUUUCUAAGCU_____UUCAA ........................................((..(((((((...(((((.............................)))))...)))))))..)).......... ( -8.71 = -7.88 + -0.83)

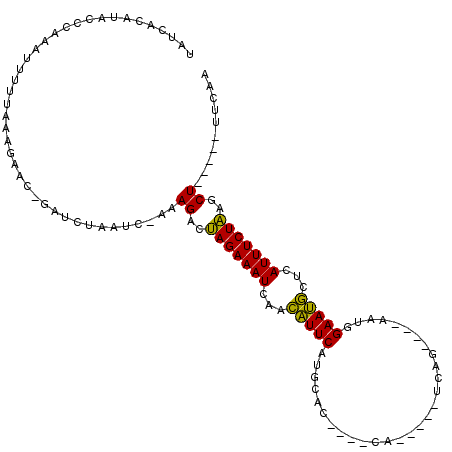
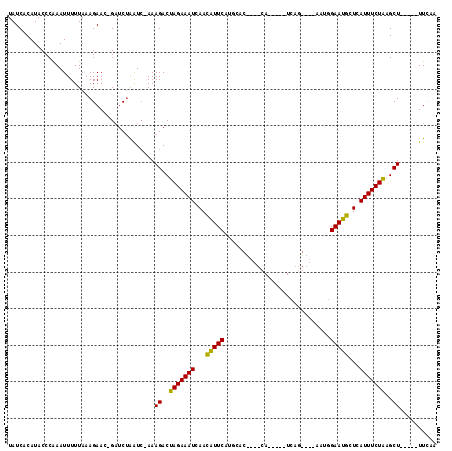
| Location | 6,042,861 – 6,042,950 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -15.90 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042861 89 - 6264404 UCGAA-----AGCUCAGAAAUGAAUGUUC-----------------------GUGAAUGAACAUUGAUUUCUGGUCUUUGCACCAGAACUGUUCUUUAAAAAUUCGGGUAUGUGAUA ...((-----((..(((((((.(((((((-----------------------(....)))))))).)))))))..))))..(((.(((..............))).)))........ ( -22.74) >pss.1919 3850729 105 - 6093698 UUGAA-----AGCUUAGAAAUGAGCAUUCCAUUGAUCCUGA-----UGAUCAGUGCAUGAAUGUUGAUUUCUAGUCUUU-GAUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUA ...((-----((.((((((((.(((((((((((((((....-----.))))))))...))))))).)))))))).))))-.((((.((.-(..(...........)..))).)))). ( -28.00) >psp.1833 3634730 105 - 5928787 UUGAA-----AGCUUAGAAAUGAGCAUUCCAUUGAUCCUGA-----UGAUCAGUGCAUGAAUGUUGAUUUCUAGUCUUU-GAUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUA ...((-----((.((((((((.(((((((((((((((....-----.))))))))...))))))).)))))))).))))-.((((.((.-(..(...........)..))).)))). ( -28.00) >pf5.3378 6386031 84 - 7074893 ---AA-----AGCUUAGAAAUGAUCAUUCACUU------------------------UGAAUGUUGAUUUCUAGUCUUUUGAUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUA ---..-----.((((((((((.(.(((((....------------------------.))))).).)))(((((((....)))))))..-............)))))))........ ( -15.40) >pfo.3002 5729650 97 - 6438405 ---AAGUAAAAGCUUAGAAAUGAGCAUUCCAUCG---UUGA-----UG--------GUGAAUGUUGAUUUCUAGUCUUUUGACUGGUUC-GUUCUUUAAAAAUUUGGGUAUGUGAUA ---.((((((((.((((((((.(((((((((((.---..))-----))--------)..)))))).)))))))).))))).))).....-........................... ( -22.80) >pst.2031 3872730 106 - 6397126 UUGAA-----AGCUUAGAAAUGAGCAUUCCAC----CCUGAGAGAUUGAAGGGUGCGUGAAUGUUGAUUUCUAGUCUUU-GAUUAGAUC-GUUCUUUAAAAAUUUGGGUAUGUGAUA ...((-----((.((((((((.((((((((((----(((..........))))))...))))))).)))))))).))))-.((((.((.-(..(...........)..))).)))). ( -28.60) >consensus UUGAA_____AGCUUAGAAAUGAGCAUUCCAUU____CUGA_____UG____GUGCAUGAAUGUUGAUUUCUAGUCUUU_GAUUAGAUC_GUUCUUUAAAAAUUUGGGUAUGUGAUA ...........((((((((((.(((((((.............................))))))).)))(((((((....)))))))...............)))))))........ (-15.90 = -15.13 + -0.77)

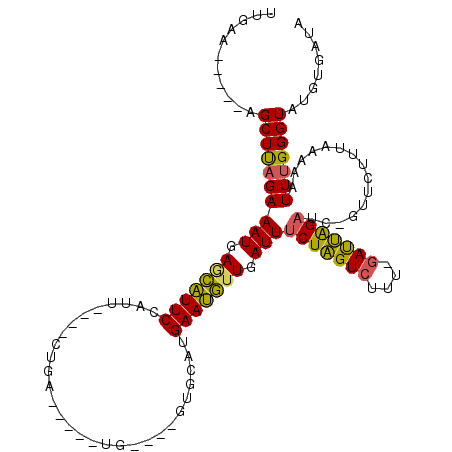
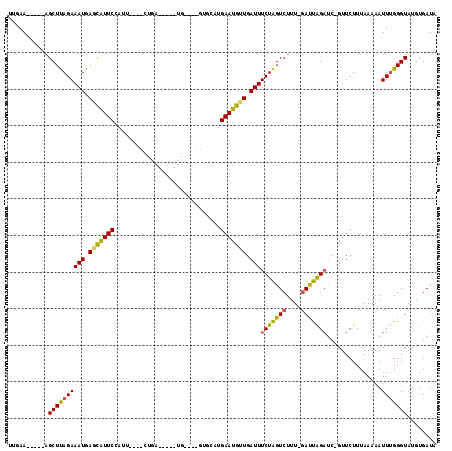
| Location | 6,042,901 – 6,042,984 |
|---|---|
| Length | 83 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.95 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -14.71 |
| Energy contribution | -13.93 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3201 6042901 83 - 6264404 CGAUCCUCCUUGGCUCCACCAUCUAAAACAAUCG--UCGAA-----AGCUCAGAAAUGAAUGUUC-----------------------GUGAAUGAACAUUGAUUUCUGGUCU ((((......(((.....))).........))))--.....-----((..(((((((.(((((((-----------------------(....)))))))).)))))))..)) ( -20.26) >pss.1919 3850767 103 - 6093698 CGAUCCCGCUUGGCUCCACCACUUACUGCUUCUGUUUUGAA-----AGCUUAGAAAUGAGCAUUCCAUUGAUCCUGA-----UGAUCAGUGCAUGAAUGUUGAUUUCUAGUCU .(((...((((((.....))..(((..((....))..))))-----))).(((((((.(((((((((((((((....-----.))))))))...))))))).)))))))))). ( -28.20) >psp.1833 3634768 102 - 5928787 CGAUCCCGCUUGGCUCCACCACUUACUGCUUCUG-UUUGAA-----AGCUUAGAAAUGAGCAUUCCAUUGAUCCUGA-----UGAUCAGUGCAUGAAUGUUGAUUUCUAGUCU ...........(((.............(((((..-...).)-----))).(((((((.(((((((((((((((....-----.))))))))...))))))).)))))))))). ( -27.90) >pf5.3378 6386070 79 - 7074893 CGAUCCCGCUUGGCUCCACCAUAAACUGCUUCUG-----AA-----AGCUUAGAAAUGAUCAUUCACUU------------------------UGAAUGUUGAUUUCUAGUCU .(((...(((((((.............)))....-----.)-----))).(((((((.(.(((((....------------------------.))))).).)))))))))). ( -15.52) >pfo.3002 5729689 91 - 6438405 CGAUCCCGCUUGGCUCCACCACUA-CUGCUUCUG-----AAGUAAAAGCUUAGAAAUGAGCAUUCCAUCG---UUGA-----UG--------GUGAAUGUUGAUUUCUAGUCU .(((...((((((.....))..((-((.......-----.)))).)))).(((((((.(((((((((((.---..))-----))--------)..)))))).)))))))))). ( -23.40) >pst.2031 3872768 103 - 6397126 CGAUCCCGCUUGGCUCCACCACUUACUGCUUCUG-UUUGAA-----AGCUUAGAAAUGAGCAUUCCAC----CCUGAGAGAUUGAAGGGUGCGUGAAUGUUGAUUUCUAGUCU ...........(((.............(((((..-...).)-----))).(((((((.((((((((((----(((..........))))))...))))))).)))))))))). ( -28.50) >consensus CGAUCCCGCUUGGCUCCACCACUUACUGCUUCUG__UUGAA_____AGCUUAGAAAUGAGCAUUCCAUU____CUGA_____UG____GUGCAUGAAUGUUGAUUUCUAGUCU ..........(((.....))).........................((.((((((((.(((((((.............................))))))).)))))))).)) (-14.71 = -13.93 + -0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:04:27 2007