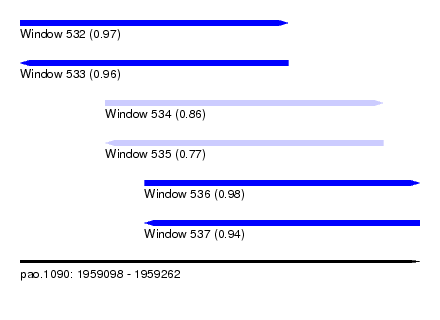
| Sequence ID | pao.1090 |
|---|---|
| Location | 1,959,098 – 1,959,262 |
| Length | 164 |
| Max. P | 0.981860 |

| Location | 1,959,098 – 1,959,208 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
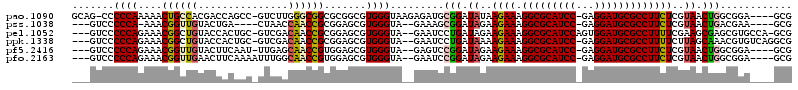
>pao.1090 1959098 110 + 6264404 CGGU---UAGAAUACCGGCCUGU-CACGCCGGGGGUCGCGGGUUCGAGUCCCGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUAUAUCCGCAUCUC .(((---......)))(((....-...))).(((((.(((((.......)))).).)))))((....((..((((((((((((....)))))))).))))..))...))..... ( -44.20) >pss.1038 1989903 89 + 6093698 CGGU----------------UGC-GACACCG------AGGGGUUCGAGCCCUA-CCGCUUCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC- ((((----------------...-...))))------.(((((....))))).-..((.((((.....))))(((((((((((....))))))))))).........))....- ( -33.70) >pf5.2416 2473580 105 - 7074893 CGGU--------CUACCACCGGUUCACGCCGAGGGUCGCGGGUUUAACCUUCGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGGACUC- .((.--------...)).((((.....((.(((((.((.(((.....))).)).)).))).)).......(((((((((((((....)))))))).)))))....))))....- ( -43.70) >pfo.2163 2257081 111 - 6438405 AGGUAGUUGAAACAGUUGCCUGU-CACACCG-GGAUCGAGGGUUCGAGCCCUGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGGAUUC- (((((..((...))..)))))..-....(((-((.(((..(((..((((.......)))).))).....)))(((((((((((....)))))))))))......)))))....- ( -42.10) >psp.1012 1967328 89 + 5928787 CGGU----------------UGC-AACACCG------AGGGGUUCGAGCCCUA-CCGCUCCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC- ((((----------------...-...))))------.(((((....))))).-..((..(((.....))).(((((((((((....))))))))))).........))....- ( -32.20) >pst.2206 2193377 89 - 6397126 CGGU----------------UGC-GACACCG------AAGGGUUCGAGCCCUA-CCGCUUCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC- .(((----------------(.(-((.(((.------...))))))))))...-..((.((((.....))))(((((((((((....))))))))))).........))....- ( -35.30) >consensus CGGU________________UGC_CACACCG______AAGGGUUCGAGCCCUA_CCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCAUUC_ (((...................................((((......))))..................(((((((((((((....)))))))).)))))....)))...... (-25.74 = -25.72 + -0.03)
| Location | 1,959,098 – 1,959,208 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1090 1959098 110 - 6264404 GAGAUGCGGAUAUAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACGGGACUCGAACCCGCGACCCCCGGCGUG-ACAGGCCGGUAUUCUA---ACCG ......(((.((..((((.((((((((....))))))))))))..))(((((((..(((..((((..(((......)))..)))).))).-.)..))))))......---.))) ( -47.30) >pss.1038 1989903 89 - 6093698 -GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGAAGCGG-UAGGGCUCGAACCCCU------CGGUGUC-GCA----------------ACCG -.....(((......((((((((((((....)))))))))).)).....((.(((.((.(-..(((......)))..------).)).))-)))----------------.))) ( -34.30) >pf5.2416 2473580 105 + 7074893 -GAGUCCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACGAAGGUUAAACCCGCGACCCUCGGCGUGAACCGGUGGUAG--------ACCG -..(.((((.((..((((.((((((((....))))))))))))..)).)))))((.((((((....)).....)))).(((.((((......)))).)))..--------.)). ( -43.40) >pfo.2163 2257081 111 + 6438405 -GAAUCCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACAGGGCUCGAACCCUCGAUCC-CGGUGUG-ACAGGCAACUGUUUCAACUACCU -...((((..(((..((((((((((((....)))))))))).))....))).))))..((...(((.((((....)))).))-)(((..(-((((....)))))...))).)). ( -47.40) >psp.1012 1967328 89 - 5928787 -GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGGAGCGG-UAGGGCUCGAACCCCU------CGGUGUU-GCA----------------ACCG -.....(((......((((((((((((....)))))))))).))(((((..((.(((.((-(.........))).))------).)))))-)).----------------.))) ( -35.20) >pst.2206 2193377 89 + 6397126 -GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGAAGCGG-UAGGGCUCGAACCCUU------CGGUGUC-GCA----------------ACCG -.....(((......((((((((((((....)))))))))).)).....((.(((.((.(-.((((......)))).------).)).))-)))----------------.))) ( -35.10) >consensus _GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGGAGCGG_CAGGGCUCGAACCCCC______CGGUGUG_ACA________________ACCG ......(((.((..((((.((((((((....))))))))))))..)).)))(((((((.......))))....((.........)))))......................... (-24.34 = -24.45 + 0.11)
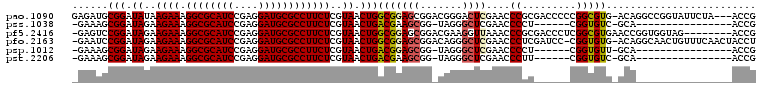
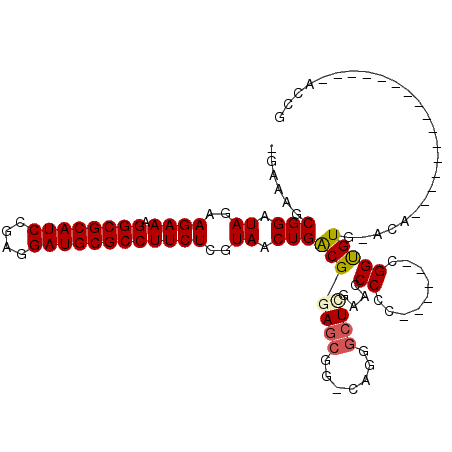
| Location | 1,959,133 – 1,959,247 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.04 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1090 1959133 114 + 6264404 GGGUUCGAGUCCCGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUAUAUCCGCAUCUCUUACCCACGCCGCGCCGCCCAAGAC-GGCUGGUCGUGGCA ((((..(((....(....)...((....((..((((((((((((....)))))))).))))..))...))..)))..))))..(((((((((((......-))).)).)))))). ( -50.10) >pss.1038 1989919 108 + 6093698 GGGUUCGAGCCCUA-CCGCUUCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC--UACCCACGCUCCGCGGUUGGUUAG----UCAGUACAACC ((((..((((....-.....((((.....))))(((((((((((....))))))))))).........))))..--.)))).........(((((.....----......))))) ( -36.90) >pf5.2416 2473611 112 - 7074893 GGGUUUAACCUUCGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGGACUC--UACCCACGCUCCACGGUUGCUCAA-AUUGAAGUACAACC ((((....((.((((..(((.....))).))))(((((((((((....))))))))))).........))....--.)))).........(((((..(..-......)..))))) ( -34.80) >pfo.2163 2257118 113 - 6438405 GGGUUCGAGCCCUGUCCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGGAUUC--UACCCACGCUCCACGGUUGCCAAAUUUUGAAGUUCAACC ((((..((((.......))))..((.(.((.(((((((((((((....)))))))).))))).)).).))....--.))))..(((.((..(((....)))..)).)))...... ( -35.80) >psp.1012 1967344 108 + 5928787 GGGUUCGAGCCCUA-CCGCUCCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC--UACCCACGCUCCGCGGUUGGUUAG----UCAGUUCAACC .((((.((((((.(-((((..(((.....))).(((((((((((....)))))))))))...............--............))))).))....----...)))))))) ( -36.01) >pst.2206 2193393 108 - 6397126 GGGUUCGAGCCCUA-CCGCUUCGUCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCUUUC--UACCCACGCUCCGCGGUUGGUUAG----UCAGUUCAACC .((((.((((((.(-((((.((((.....))))(((((((((((....)))))))))))...............--............))))).))....----...)))))))) ( -37.51) >consensus GGGUUCGAGCCCUA_CCGCUCCGCCAGUUACGAGAAGGCGCAUCCUCGGAUGCGCCUUUCUUCUAUCCGCAUUC__UACCCACGCUCCGCGGUUGGUUAG____UCAGUUCAACC ((((..((((.......))))..........(((((((((((((....)))))))).)))))...............)))).........(((((...............))))) (-30.24 = -30.04 + -0.19)

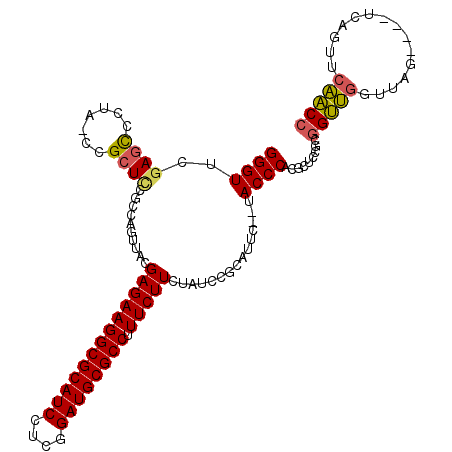
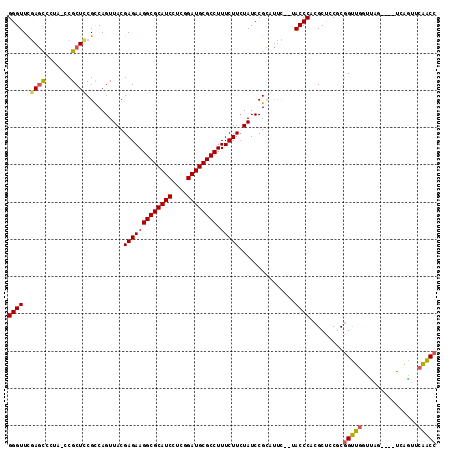
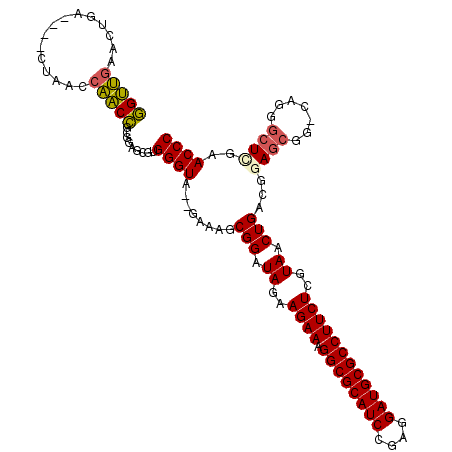
| Location | 1,959,133 – 1,959,247 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -41.99 |
| Consensus MFE | -33.66 |
| Energy contribution | -33.58 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1090 1959133 114 - 6264404 UGCCACGACCAGCC-GUCUUGGGCGGCGCGGCGUGGGUAAGAGAUGCGGAUAUAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACGGGACUCGAACCC ..(((((.((.(((-((.....)))))..)))))))((..(...(((((.((..((((.((((((((....))))))))))))..)).)).)))...)..))(((.......))) ( -50.90) >pss.1038 1989919 108 - 6093698 GGUUGUACUGA----CUAACCAACCGCGGAGCGUGGGUA--GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGAAGCGG-UAGGGCUCGAACCC ((((((....)----).)))).(((((....(((.(((.--....(((......((((.((((((((....))))))))))))))).))).)))..))))-).(((......))) ( -40.10) >pf5.2416 2473611 112 + 7074893 GGUUGUACUUCAAU-UUGAGCAACCGUGGAGCGUGGGUA--GAGUCCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACGAAGGUUAAACCC ((((((.(......-..).)))))).........((((.--...((((..(((..((((((((((((....)))))))))).))....))).))))...(((....)))..)))) ( -41.40) >pfo.2163 2257118 113 + 6438405 GGUUGAACUUCAAAAUUUGGCAACCGUGGAGCGUGGGUA--GAAUCCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGGCGGAGCGGACAGGGCUCGAACCC (((((...............)))))...((((.(..((.--...((((..(((..((((((((((((....)))))))))).))....))).))))....)).).))))...... ( -40.26) >psp.1012 1967344 108 - 5928787 GGUUGAACUGA----CUAACCAACCGCGGAGCGUGGGUA--GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGGAGCGG-UAGGGCUCGAACCC (((((......----.))))).(((((....(((.(((.--....(((......((((.((((((((....))))))))))))))).))).)))..))))-).(((......))) ( -39.10) >pst.2206 2193393 108 + 6397126 GGUUGAACUGA----CUAACCAACCGCGGAGCGUGGGUA--GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGAAGCGG-UAGGGCUCGAACCC (((((......----.))))).(((((....(((.(((.--....(((......((((.((((((((....))))))))))))))).))).)))..))))-).(((......))) ( -40.20) >consensus GGUUGAACUGA____CUAACCAACCGCGGAGCGUGGGUA__GAAAGCGGAUAGAAGAAAGGCGCAUCCGAGGAUGCGCCUUCUCGUAACUGACGGAGCGG_CAGGGCUCGAACCC (((((...............))))).........((((........(((.((..((((.((((((((....))))))))))))..)).)))...((((.......))))..)))) (-33.66 = -33.58 + -0.08)

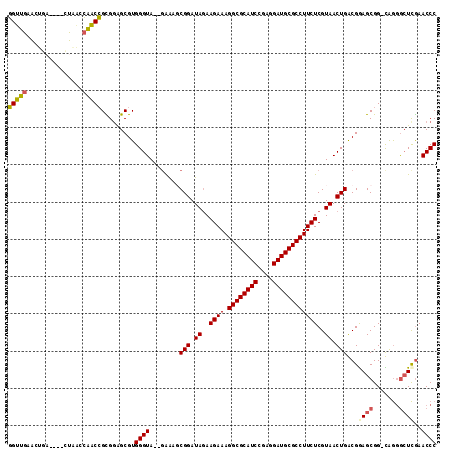
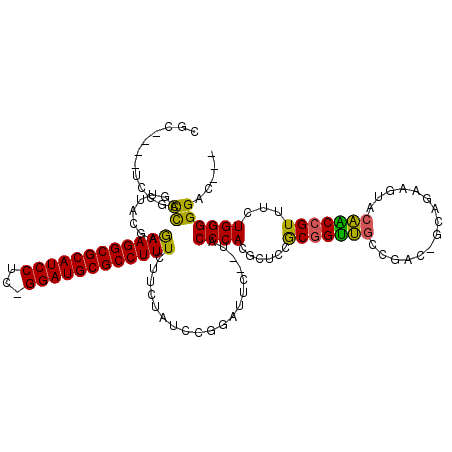
| Location | 1,959,149 – 1,959,262 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -32.98 |
| Energy contribution | -31.76 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1090 1959149 113 + 6264404 CGC----UCCGCCAGUUACGAGAAGGCGCAUCCUC-GGAUGCGCCUUUCUUAUAUCCGCAUCUCUUACCCACGCCGCGCCGCCCAAGAC-GGCUGGUCGUGGCAGUUUUUGGGGG-CUGC .((----.((.((((..(((((((((((((((...-.)))))))).))))).....................(((((((((((......-))).)).)))))).))..)))))))-)... ( -51.50) >pss.1038 1989934 105 + 6093698 CGC----UUCGUCAGUUACGAGAAGGCGCAUCCUC-GGAUGCGCCUUUCUUCUAUCCGCUUUC--UACCCACGCUCCGCGGUUGGUUAG----UCAGUACAACCGUUU-UGGGGGAC--- .((----.((((.....))))(((((((((((...-.))))))))))).........))..((--(.((((......(((((((.....----......)))))))..-))))))).--- ( -37.90) >pel.1052 1961730 113 + 5888780 CGC-UGGCACGCUCGCUUCGAAAAGGCGCAUCCACUGGAUGCGCCUUUCUUCUAUCAGGAUUC--UACCCACGCUCCGCGGUUGUCGAC-GCAGUGGUACAGCCGUUUCUGGGGGAC--- .((-((...((((.((.((((((((((((((((...)))))))))))).........((....--...)).(((...)))....)))).-))))))...)))).((((.....))))--- ( -40.80) >ppk.1338 2633420 113 + 6181863 CGCCUGACACGUUUGCUAAGAAAAGGCGCAUCCUC-GGAUGCGCCUUUCUUUUAUCAGGAUUC--UACCCACGCUCCGCGGUUGUCGAC-GCAGUGGUACAGCCGUUUCUGGGGGAC--- ..(((((.........((((((((((((((((...-.))))))))))).))))))))))..((--(.((((......(((((((((.((-...)).).))))))))...))))))).--- ( -43.40) >pf5.2416 2473627 109 - 7074893 CGC----UCCGCCAGUUACGAGAAGGCGCAUCCUC-GGAUGCGCCUUUCUUCUAUCCGGACUC--UACCCACGCUCCACGGUUGCUCAA-AUUGAAGUACAACCGUUUCUGGGGGAC--- ...----(((.((((..(((((((((((((((...-.))))))))))..........((....--...))...)))...(((((..(..-......)..)))))))..)))).))).--- ( -42.10) >pfo.2163 2257134 110 - 6438405 CGC----UCCGCCAGUUACGAGAAGGCGCAUCCUC-GGAUGCGCCUUUCUUCUAUCCGGAUUC--UACCCACGCUCCACGGUUGCCAAAUUUUGAAGUUCAACCGUUUCUGGGGGAC--- ...----(((.((((..(((((((((((((((...-.))))))))))..........((....--...))...)))...(((((...((((....)))))))))))..)))).))).--- ( -40.30) >consensus CGC____UCCGCCAGUUACGAGAAGGCGCAUCCUC_GGAUGCGCCUUUCUUCUAUCCGGAUUC__UACCCACGCUCCGCGGUUGCCGAC_GCAGAAGUACAACCGUUUCUGGGGGAC___ ...........((........((((((((((((...))))))))))))...................((((......(((((((...............)))))))...))))))..... (-32.98 = -31.76 + -1.22)
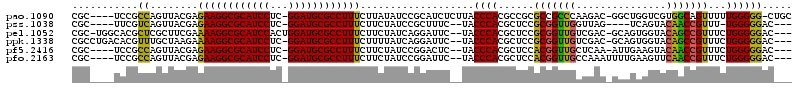

| Location | 1,959,149 – 1,959,262 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -30.29 |
| Energy contribution | -29.88 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
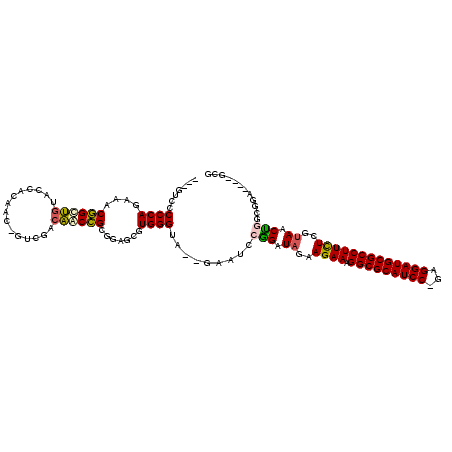
>pao.1090 1959149 113 - 6264404 GCAG-CCCCCAAAAACUGCCACGACCAGCC-GUCUUGGGCGGCGCGGCGUGGGUAAGAGAUGCGGAUAUAAGAAAGGCGCAUCC-GAGGAUGCGCCUUCUCGUAACUGGCGGA----GCG ...(-(((((........(((((.((.(((-((.....)))))..)))))))...........((.((..((((.((((((((.-...))))))))))))..)).)))).)).----)). ( -51.50) >pss.1038 1989934 105 - 6093698 ---GUCCCCCA-AAACGGUUGUACUGA----CUAACCAACCGCGGAGCGUGGGUA--GAAAGCGGAUAGAAGAAAGGCGCAUCC-GAGGAUGCGCCUUCUCGUAACUGACGAA----GCG ---((((....-....((((((....)----).))))..(((((...)))))...--......))))....(((.((((((((.-...)))))))))))((((.....)))).----... ( -34.20) >pel.1052 1961730 113 - 5888780 ---GUCCCCCAGAAACGGCUGUACCACUGC-GUCGACAACCGCGGAGCGUGGGUA--GAAUCCUGAUAGAAGAAAGGCGCAUCCAGUGGAUGCGCCUUUUCGAAGCGAGCGUGCCA-GCG ---..............(((((((...(((-.((((...(((((...)))))...--...............((((((((((((...)))))))))))))))).)))...))).))-)). ( -40.30) >ppk.1338 2633420 113 - 6181863 ---GUCCCCCAGAAACGGCUGUACCACUGC-GUCGACAACCGCGGAGCGUGGGUA--GAAUCCUGAUAAAAGAAAGGCGCAUCC-GAGGAUGCGCCUUUUCUUAGCAAACGUGUCAGGCG ---.....((.((.(((..(((.((((.((-..((.(....)))..))))))...--............((((((((((((((.-...)))))))).)))))).)))..))).)).)).. ( -40.70) >pf5.2416 2473627 109 + 7074893 ---GUCCCCCAGAAACGGUUGUACUUCAAU-UUGAGCAACCGUGGAGCGUGGGUA--GAGUCCGGAUAGAAGAAAGGCGCAUCC-GAGGAUGCGCCUUCUCGUAACUGGCGGA----GCG ---.((((((((..((((((((.(......-..).))))))))....).))))..--..(.((((.((..((((.((((((((.-...))))))))))))..)).))))))))----... ( -46.00) >pfo.2163 2257134 110 + 6438405 ---GUCCCCCAGAAACGGUUGAACUUCAAAAUUUGGCAACCGUGGAGCGUGGGUA--GAAUCCGGAUAGAAGAAAGGCGCAUCC-GAGGAUGCGCCUUCUCGUAACUGGCGGA----GCG ---.((((((((..(((((((...............)))))))....).))))..--....((((.((..((((.((((((((.-...))))))))))))..)).)))).)))----... ( -41.56) >consensus ___GUCCCCCAGAAACGGCUGUACCACAAC_GUCGACAACCGCGGAGCGUGGGUA__GAAUCCGGAUAGAAGAAAGGCGCAUCC_GAGGAUGCGCCUUCUCGUAACUGGCGGA____GCG .......((((....((((((...............)))))).......)))).........(((.((..((((.(((((((((...)))))))))))))..)).)))............ (-30.29 = -29.88 + -0.41)
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:13:01 2007