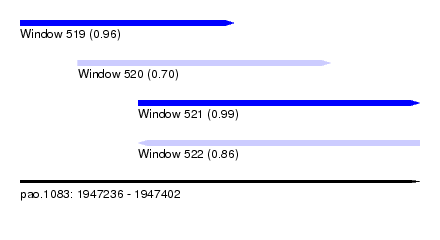
| Sequence ID | pao.1083 |
|---|---|
| Location | 1,947,236 – 1,947,402 |
| Length | 166 |
| Max. P | 0.989126 |

| Location | 1,947,236 – 1,947,325 |
|---|---|
| Length | 89 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 61.86 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -9.05 |
| Energy contribution | -8.50 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
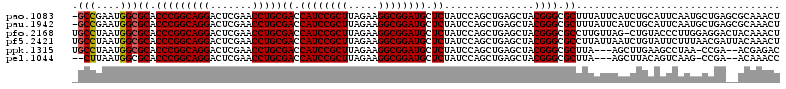
>pao.1083 1947236 89 + 6264404 UAAG----------------AGGUAAUGCAAGUCGGUUGCUGACU------UUGUGAU----UGCCAGAGCUUAAAGUUUGCGCUCAGCAUUGAAUGCAGAUGAAUAAAGCGCCC ....----------------.(((((((((((((((...))))).------)))).))----))))...((((...(((((((.((......)).))))))).....)))).... ( -26.10) >pst.2216 2180417 105 - 6397126 UAAGCUAGCACUGGUUGAUUAGGUCAGGCAGA---GUUGCAAAUCAGCUCGGUGUGAU----UGAAAGCCCUUU-AGUUUGUUAUCG--CAAGAAUACAAAUUAUAUAAGUGCCC .......(((((((((.....(((((.((.((---((((.....)))))).)).))))----)...))))...(-(((((((..((.--...))..))))))))....))))).. ( -33.70) >psp.1004 1952461 105 + 5928787 UAAGCUAGCACUGGUUGAUUAGGUCAGGCAGA---GUUGCAAAUCAGCUCGGUGUGAU----UGAAAGCCUUUU-AGUUUGUUAUCG--CAAGAAUACAAAUUAUAUAAGUGCCC .......(((((((((.....(((((.((.((---((((.....)))))).)).))))----)...))))...(-(((((((..((.--...))..))))))))....))))).. ( -34.10) >pfo.2168 2249639 86 - 6438405 UAAGA---------------AGGUCGAGCGAA---AUCAACGUUU----UGCAAGGCU----GGAAAGCACUU--AGUUUGUAGUCCUCCAAGGGUACAG-CUAACAAGGCGCCC .....---------------.((((..(((((---((....))))----)))..))))----((...((..((--((((.(((.(((.....))))))))-))))....)).)). ( -22.20) >pf5.2421 2466274 91 - 7074893 UAAGA---------------AGGUCGGGCAGA---GUUGAUGUUUAACUCGUAACGAC----UGAAAGCAAUU--AGUUUGUAAUCGUUAAAGAAUACAGAUUAAUAAGGCGCCC .....---------------.....((((.((---(((((...)))))))........----.....((.(((--((((((((.((......)).)))))))))))...)))))) ( -26.60) >pss.1031 1981257 109 + 6093698 UAAGCUAGCACUGGUUGAUUCGGUCAGGCAGA---GUUGCAAAUCAGCUCGGUGUGAUCGAUUGAAAGCCUUUU-AGUUUGUUAUCG--CAAGAGUACAAAUUAUAUAAGUGCCC .......(((((((((...(((((((.((.((---((((.....)))))).)).))))))).....))))...(-(((((((..((.--...))..))))))))....))))).. ( -39.90) >consensus UAAGC_______________AGGUCAGGCAGA___GUUGCAAAUCAGCUCGGUGUGAU____UGAAAGCCCUUU_AGUUUGUUAUCG__CAAGAAUACAAAUUAAAAAAGCGCCC .....................(((...((((.....))))...........................((......(((((((..((......))..)))))))......))))). ( -9.05 = -8.50 + -0.55)

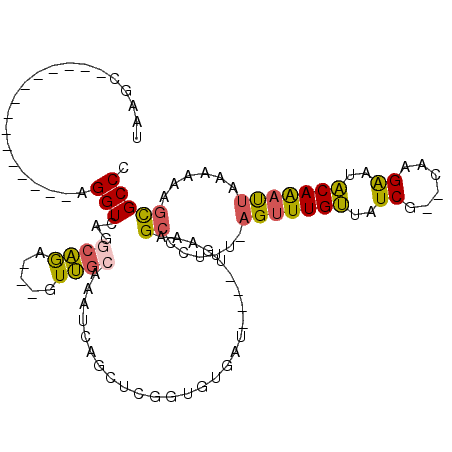
| Location | 1,947,260 – 1,947,365 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.07 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -20.87 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1083 1947260 105 + 6264404 UGACU------UUGUGAUUGCCAGAGCUUAAAGUUUGCGCUCAGCAUUGAAUGCAGAUGAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG ...((------((((.....((((.((((...(((((((.((......)).))))))).....))))((.....))....))))))))))(((((((.......))))))) ( -29.30) >pau.1942 2850233 105 - 6537648 UGACU------UUGUGAUUGCCAGAGCUUAAAGUUUGCGCUCAGCAUUGAAUGCAGAUGAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG ...((------((((.....((((.((((...(((((((.((......)).))))))).....))))((.....))....))))))))))(((((((.......))))))) ( -29.30) >pst.2216 2180454 108 - 6397126 AAAUCAGCUCGGUGUGAUUGAAAGCCCUUU-AGUUUGUUAUCG--CAAGAAUACAAAUUAUAUAAGUGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG ...((((((.(((.....((...((.((((-(((((((..((.--...))..))))))))...))).)).))..))).))))))......(((((((.......))))))) ( -30.30) >psp.1004 1952498 108 + 5928787 AAAUCAGCUCGGUGUGAUUGAAAGCCUUUU-AGUUUGUUAUCG--CAAGAAUACAAAUUAUAUAAGUGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG ...((((((.(((.....((...(((((.(-(((((((..((.--...))..))))))))...))).)).))..))).))))))......(((((((.......))))))) ( -31.80) >pfo.2168 2249661 104 - 6438405 CGUUU----UGCAAGGCUGGAAAGCACUU--AGUUUGUAGUCCUCCAAGGGUACAG-CUAACAAGGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG .((((----((...((((((...((..((--((((.(((.(((.....))))))))-))))....))((.....))))))))...))))))((((((.......)))))). ( -31.60) >pf5.2421 2466296 109 - 7074893 UGUUUAACUCGUAACGACUGAAAGCAAUU--AGUUUGUAAUCGUUAAAGAAUACAGAUUAAUAAGGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG .......(((....((.((((..((.(((--((((((((.((......)).)))))))))))...))((.....)))))).))....)))(((((((.......))))))) ( -31.60) >consensus UGAUU____CGUUGUGAUUGAAAGCACUU__AGUUUGUAAUCG_CAAAGAAUACAGAUUAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUG .......................((.(((..((((((((.((......)).))))))))....))).)).....((((.........))))((((((.......)))))). (-20.87 = -20.82 + -0.05)

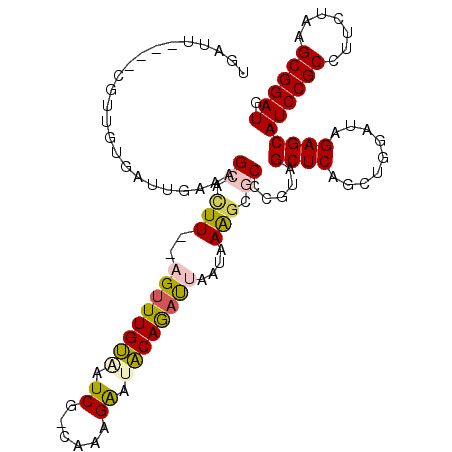
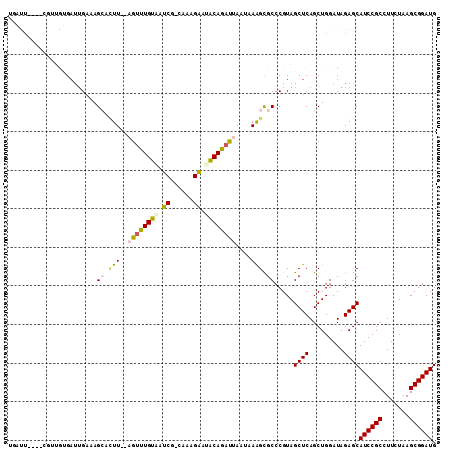
| Location | 1,947,285 – 1,947,402 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -48.08 |
| Consensus MFE | -44.83 |
| Energy contribution | -44.70 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
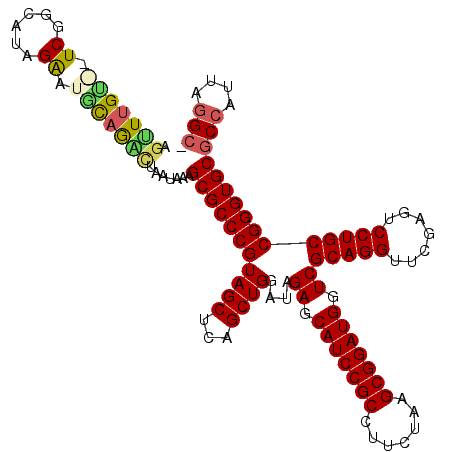
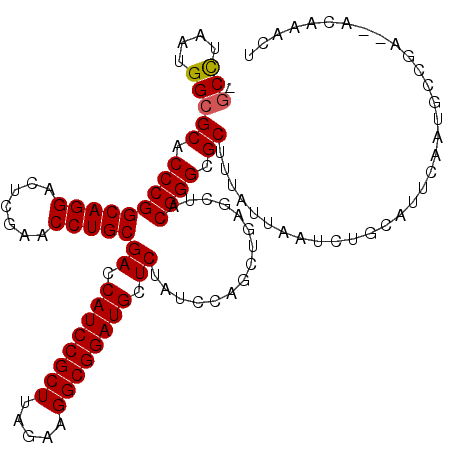
>pao.1083 1947285 117 + 6264404 AGUUUGCGCUCAGCAUUGAAUGCAGAUGAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUCGGC- .(((((((.((......)).))))))).......(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....)))- ( -48.80) >pau.1942 2850258 117 - 6537648 AGUUUGCGCUCAGCAUUGAAUGCAGAUGAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUCGGC- .(((((((.((......)).))))))).......(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....)))- ( -48.80) >pfo.2168 2249686 117 - 6438405 AGUUUGUAGUCCUCCAAGGGUACAG-CUAACAAGGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUAGGCA .(((.((.(((((....))).)).)-).)))...(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....))). ( -48.60) >pf5.2421 2466325 118 - 7074893 AGUUUGUAAUCGUUAAAGAAUACAGAUUAAUAAGGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUAGGCA ((((((((.((......)).))))))))......(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....))). ( -49.70) >ppk.1315 2585732 112 + 6181863 GUCUCGU--UCGG-UUAGGCUUCAAGCU---UAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUAGGCA .......--....-((((((.....)))---)))(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....))). ( -47.60) >pel.1044 1952614 110 + 5888780 GGUUUGU--UCGG-CUUGACUGUAAGCU---UAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUAAG-- (((....--..((-((((....))))))---...(((((((((((...))))....((.(((((((.......))))))).))(((((.......)))))))))))))))......-- ( -45.00) >consensus AGUUUGU__UCGGCAUAGAAUGCAGACUAAUAAAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUAGGC_ .(((((((.((......)).))))))).......(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))(((....))). (-44.83 = -44.70 + -0.13)
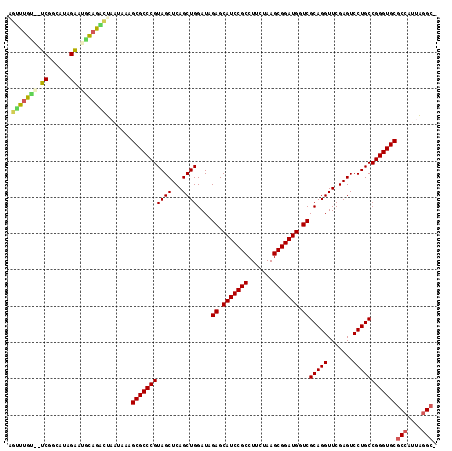
| Location | 1,947,285 – 1,947,402 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.93 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.1083 1947285 117 - 6264404 -GCCGAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUUUAUUCAUCUGCAUUCAAUGCUGAGCGCAAACU -(((((((((.((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).)))))).((.((((...)))).))).))..... ( -44.10) >pau.1942 2850258 117 + 6537648 -GCCGAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUUUAUUCAUCUGCAUUCAAUGCUGAGCGCAAACU -(((((((((.((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).)))))).((.((((...)))).))).))..... ( -44.10) >pfo.2168 2249686 117 + 6438405 UGCCUAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCCUUGUUAG-CUGUACCCUUGGAGGACUACAAACU .((.(((..(.((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).)..))))-)((((.((.....))..)))).... ( -41.30) >pf5.2421 2466325 118 + 7074893 UGCCUAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCCUUAUUAAUCUGUAUUCUUUAACGAUUACAAACU .(((....)))((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).))..........((((.((......)).)))).... ( -41.50) >ppk.1315 2585732 112 - 6181863 UGCCUAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUUA---AGCUUGAAGCCUAA-CCGA--ACGAGAC .(((....)))((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).))(((---.((.....)).)))-....--....... ( -39.40) >pel.1044 1952614 110 - 5888780 --CUUAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUUA---AGCUUACAGUCAAG-CCGA--ACAAACC --.....((((((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).))...---.((.....))...)-))).--....... ( -38.40) >consensus _GCCUAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUUUAUUAAUCUGCAUUCAAUGCCGA__ACAAACU .(((....)))((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).................................. (-36.90 = -36.93 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:12:45 2007