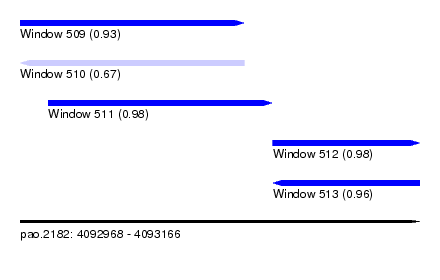
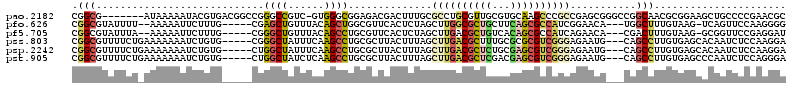
| Sequence ID | pao.2182 |
|---|---|
| Location | 4,092,968 – 4,093,166 |
| Length | 198 |
| Max. P | 0.981994 |

| Location | 4,092,968 – 4,093,079 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 63.77 |
| Mean single sequence MFE | -38.21 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.45 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.62 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2182 4092968 111 + 6264404 GACCUUUUCUCCUGCGGCG-------AUAAAAAUACGUGACGGCCGGGCCGUC-GUGGGCGGAGACGACUUUGCGCCUGCGUUGCGUGCAAGCCCGCCGAGCGGGCCGGCAACGCGGAA ...........((((((((-------((..........((((((...))))))-(..((((.(((....))).))))..)))))).(((..((((((...))))))..))).))))).. ( -51.60) >pfo.626 5161350 103 - 6438405 -----UCUCUCCUGCGGCGUAUUUU--AAAAAUUCUUUG-----CGAGCUGUUUACAGCUGGCGUUCACUCUAGCUUGGCGCUGCUUCAGCGCCAUCGGAACA---UGGCUUUGUAAG- -----........(((((((.....--..........((-----(.(((((....))))).)))..............)))))))..(((.(((((......)---)))).)))....- ( -31.35) >pf5.705 5724979 103 - 7074893 -----UCUCUCCUGCGGCGUAUUUA--AAAAAUUCUUUG-----CGGGCUGUUUACAGCCUGCGUUCACUCUAGCUUGACGCUGUCACAGCGCCAUCAGAACA---CGACUUUGUAAG- -----......(((.(((((.....--...........(-----(((((((....))))))))........((((.....)))).....)))))..)))....---............- ( -29.20) >pel.2413 4501526 103 + 5888780 -----UCUCUCCUGCGGCGUACUUUUAGAAAAUUCCGUG-----CGGGCUGUUUGCAGCCUGC--UAACUGUAGCGUGGCACUGCUUCAGUGCCAGCAGCCUU---GGGCUUUAUAAA- -----.....((((.(((....................(-----(((((((....))))))))--........((.((((((((...)))))))))).))).)---))).........- ( -39.30) >pss.803 4568485 106 - 6093698 -----UCUCUCCUGCGGCGUUUUCUGAAAAAAAUCUGUG-----CGGGCUAUUUCAAGCCUGCGCUUACUUUAGCUUGACGCUUUGCGCGCGUCGGGAGAAUG---CAGCCUUGUGAGC -----.(((.(..((.(((((((((...........(((-----((((((......)))))))))...........((((((.......))))))))))))))---).))...).))). ( -39.20) >psp.2242 4393690 106 + 5928787 -----UCUCUCCUGCGGCGUUUUCUGAAAAAAAUCUGUG-----CUGGCUAUUUCAAGCCUGCGCUUACUUUAGCUUGACGCUCUGCGAGCGUCGGGAGAAUG---CAGCCUUGUGAGC -----.(((.(..((.(((((((((...........(((-----(.((((......)))).))))...........((((((((...))))))))))))))))---).))...).))). ( -38.60) >consensus _____UCUCUCCUGCGGCGUAUUUU_AAAAAAAUCUGUG_____CGGGCUGUUUACAGCCUGCGCUCACUUUAGCUUGACGCUGCGUCAGCGCCAGCAGAACG___CGGCUUUGUAAG_ ...............(((............................(((((....)))))................((((((((...)))))))).............)))........ (-15.61 = -15.45 + -0.16)


| Location | 4,092,968 – 4,093,079 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 63.77 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.05 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2182 4092968 111 - 6264404 UUCCGCGUUGCCGGCCCGCUCGGCGGGCUUGCACGCAACGCAGGCGCAAAGUCGUCUCCGCCCAC-GACGGCCCGGCCGUCACGUAUUUUUAU-------CGCCGCAGGAGAAAAGGUC ((((((((.((.(((((((...))))))).))))))...((.((((.((((((((....(....)-((((((...))))))))).)))))...-------)))))).))))........ ( -46.20) >pfo.626 5161350 103 + 6438405 -CUUACAAAGCCA---UGUUCCGAUGGCGCUGAAGCAGCGCCAAGCUAGAGUGAACGCCAGCUGUAAACAGCUCG-----CAAAGAAUUUUU--AAAAUACGCCGCAGGAGAGA----- -............---..((((..((((((((...)))))))).((....(....)((.(((((....))))).)-----)...........--..........)).))))...----- ( -30.00) >pf5.705 5724979 103 + 7074893 -CUUACAAAGUCG---UGUUCUGAUGGCGCUGUGACAGCGUCAAGCUAGAGUGAACGCAGGCUGUAAACAGCCCG-----CAAAGAAUUUUU--UAAAUACGCCGCAGGAGAGA----- -(((.....(.((---(((((((.((((((((...))))))))...))))......((.(((((....))))).)-----)...........--...))))).)....)))...----- ( -29.50) >pel.2413 4501526 103 - 5888780 -UUUAUAAAGCCC---AAGGCUGCUGGCACUGAAGCAGUGCCACGCUACAGUUA--GCAGGCUGCAAACAGCCCG-----CACGGAAUUUUCUAAAAGUACGCCGCAGGAGAGA----- -.........((.---..(((.((((((((((...)))))))).))(((.....--((.(((((....))))).)-----)..(((....)))....))).)))...)).....----- ( -33.70) >pss.803 4568485 106 + 6093698 GCUCACAAGGCUG---CAUUCUCCCGACGCGCGCAAAGCGUCAAGCUAAAGUAAGCGCAGGCUUGAAAUAGCCCG-----CACAGAUUUUUUUCAGAAAACGCCGCAGGAGAGA----- .(((.((((.(((---(........(((((.......)))))..(((......))))))).))))......((.(-----(...(.(((((....))))))...)).)).))).----- ( -32.20) >psp.2242 4393690 106 - 5928787 GCUCACAAGGCUG---CAUUCUCCCGACGCUCGCAGAGCGUCAAGCUAAAGUAAGCGCAGGCUUGAAAUAGCCAG-----CACAGAUUUUUUUCAGAAAACGCCGCAGGAGAGA----- .(((.((((.(((---(........(((((((...)))))))..(((......))))))).))))......((.(-----(...(.(((((....))))))...)).)).))).----- ( -34.00) >consensus _CUCACAAAGCCG___CAUUCUGCUGGCGCUCACACAGCGCCAAGCUAAAGUAAACGCAGGCUGCAAACAGCCCG_____CACAGAAUUUUUU_AAAAUACGCCGCAGGAGAGA_____ .(((.(...((..............(((((((...))))))).................((((......))))...............................))..).)))...... (-15.29 = -14.05 + -1.24)

| Location | 4,092,982 – 4,093,093 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 64.12 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -16.53 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2182 4092982 111 + 6264404 CGGCG-------AUAAAAAUACGUGACGGCCGGGCCGUC-GUGGGCGGAGACGACUUUGCGCCUGCGUUGCGUGCAAGCCCGCCGAGCGGGCCGGCAACGCGGAAGCUGCCCCGAACGC .((((-------..........((....(((((.(((((-(((((((....)....(((((((......).))))))))))).))).))).))))).))((....))))))........ ( -51.40) >pfo.626 5161359 108 - 6438405 CGGCGUAUUUU--AAAAAUUCUUUG-----CGAGCUGUUUACAGCUGGCGUUCACUCUAGCUUGGCGCUGCUUCAGCGCCAUCGGAACA---UGGCUUUGUAAG-UCAGUUCCAAGGGG ...........--...........(-----(.(((((....))))).)).....((((....((((((((...))))))))..(((((.---(((((.....))-)))))))).)))). ( -40.90) >pf5.705 5724988 108 - 7074893 CGGCGUAUUUA--AAAAAUUCUUUG-----CGGGCUGUUUACAGCCUGCGUUCACUCUAGCUUGACGCUGUCACAGCGCCAUCAGAACA---CGACUUUGUAAG-GCGGUUCCGAGGAU .(((((.....--...........(-----(((((((....))))))))........((((.....)))).....))))).((.((((.---((.(((....))-))))))).)).... ( -34.80) >pss.803 4568494 111 - 6093698 CGGCGUUUUCUGAAAAAAAUCUGUG-----CGGGCUAUUUCAAGCCUGCGCUUACUUUAGCUUGACGCUUUGCGCGCGUCGGGAGAAUG---CAGCCUUGUGAGCACAAUCUCCAAGGA ..(((((((((...........(((-----((((((......)))))))))...........((((((.......))))))))))))))---)..(((((.(((......)))))))). ( -43.70) >psp.2242 4393699 111 + 5928787 CGGCGUUUUCUGAAAAAAAUCUGUG-----CUGGCUAUUUCAAGCCUGCGCUUACUUUAGCUUGACGCUCUGCGAGCGUCGGGAGAAUG---CAGCCUUGUGAGCACAAUCUCCAAGGA ..(((((((((...........(((-----(.((((......)))).))))...........((((((((...))))))))))))))))---)..(((((.(((......)))))))). ( -43.10) >pst.905 4701582 111 - 6397126 CGGCGUUUUCUGAAAAAAAUCUGUG-----CUGGCUAUCUCAAGCCUGCGCUUACUUUAGCUUGACGCUCGACGAGCGUCGGGAGAAUG---CAGCCUUGUGAGCCCAAUCUCCAGGGA ..(((((((((...........(((-----(.((((......)))).))))...........((((((((...))))))))))))))))---)..(((((.(((......)))))))). ( -42.80) >consensus CGGCGUUUUCU_AAAAAAAUCUGUG_____CGGGCUAUUUCAAGCCUGCGCUUACUUUAGCUUGACGCUGUGCGAGCGCCGGCAGAACG___CAGCCUUGUGAGCACAAUCCCCAAGGA .(((............................((((......))))..............((((((((((...))))))))))...........)))...................... (-16.53 = -16.73 + 0.20)
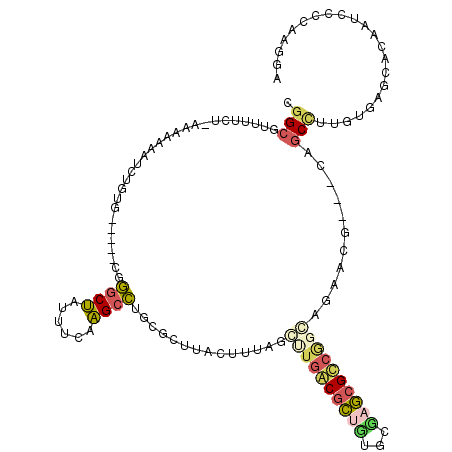
| Location | 4,093,093 – 4,093,166 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -22.85 |
| Energy contribution | -24.88 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
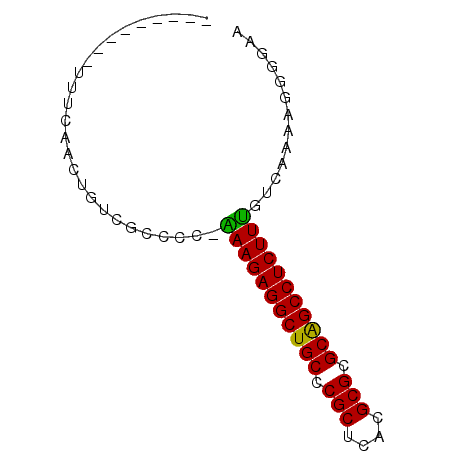
>pao.2182 4093093 73 + 6264404 UUCCCCAUUUGACGAAGAGGCCGCACGCAUCAGCGGGCAGCCUCUUCCGGGGCGACAGUCGAAACCGUCUUUG ..........(((((((((((.((.(((....))).)).)))))))).((..((.....))...))))).... ( -29.30) >pss.803 4568614 63 - 6093698 UUUCCCUUUCGACAAAGAGGCCGCACGCGUGAGCGGGCAGCCUCUUU-GGGGCGACAGUUGAAA--------- (((..((.(((.(((((((((.((.(((....))).)).))))))))-)...))).))..))).--------- ( -31.80) >ppk.898 4394766 57 - 6181863 UCCCCGUUUUGACAAAGAGGCUGCGCGCGUGAGCGGGCAGCCUCUUU-GGGGCGACAG--------------- .((((((....))(((((((((((.(((....))).)))))))))))-))))......--------------- ( -36.90) >pf5.705 5725112 62 - 7074893 CUGCCCAUUUGA-AAAGAGGCUGCGCGCGUGAGCGGGCAGCCUCUUC-UGGGCGACAGUUGAAA--------- .((((((...((-..(((((((((.(((....))).)))))))))))-))))))..........--------- ( -36.80) >pfo.626 5161486 62 - 6438405 AACCCUAUCUGA-AAAGAGGCUGCGCGCGUGAGCGGGCAGCCUCUUU-CGGGCGACAGUUGAAA--------- .......(((((-((.((((((((.(((....))).)))))))))))-))))............--------- ( -33.70) >psp.2242 4393819 63 + 5928787 UUUCUCUUUCGACAAAGAGGCCGCACGCAUGAGCGGGCAGCCUCUUU-GGGGCGACAGUUGAAA--------- .(((.((.(((.(((((((((.((.(((....))).)).))))))))-)...))).))..))).--------- ( -28.80) >consensus UUCCCCAUUUGACAAAGAGGCCGCACGCGUGAGCGGGCAGCCUCUUU_GGGGCGACAGUUGAAA_________ ...((((......((((((((.((.(((....))).)).)))))))).))))..................... (-22.85 = -24.88 + 2.03)

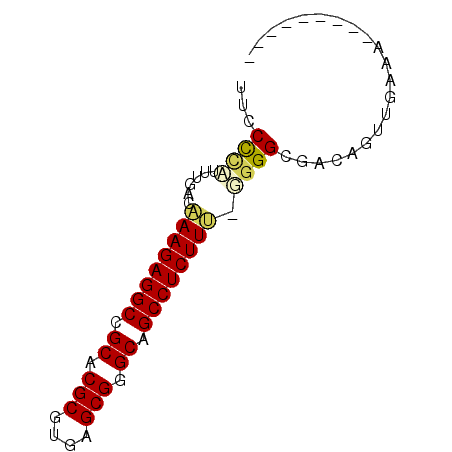
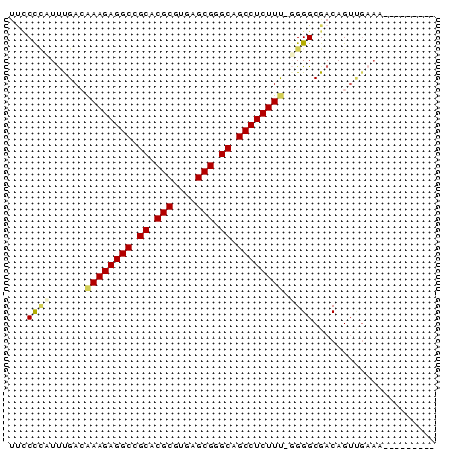
| Location | 4,093,093 – 4,093,166 |
|---|---|
| Length | 73 |
| Sequences | 6 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -24.79 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -5.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2182 4093093 73 - 6264404 CAAAGACGGUUUCGACUGUCGCCCCGGAAGAGGCUGCCCGCUGAUGCGUGCGGCCUCUUCGUCAAAUGGGGAA ....((((((....)))))).((((((((((((((((.(((....))).)))))))))))......))))).. ( -40.00) >pss.803 4568614 63 + 6093698 ---------UUUCAACUGUCGCCCC-AAAGAGGCUGCCCGCUCACGCGUGCGGCCUCUUUGUCGAAAGGGAAA ---------.(((..((.(((...(-(((((((((((.(((....))).)))))))))))).))).)).))). ( -29.80) >ppk.898 4394766 57 + 6181863 ---------------CUGUCGCCCC-AAAGAGGCUGCCCGCUCACGCGCGCAGCCUCUUUGUCAAAACGGGGA ---------------......((((-(((((((((((.(((....))).)))))))))))((....)))))). ( -31.40) >pf5.705 5725112 62 + 7074893 ---------UUUCAACUGUCGCCCA-GAAGAGGCUGCCCGCUCACGCGCGCAGCCUCUUU-UCAAAUGGGCAG ---------...........(((((-(((((((((((.(((....))).)))))))))))-.....))))).. ( -33.30) >pfo.626 5161486 62 + 6438405 ---------UUUCAACUGUCGCCCG-AAAGAGGCUGCCCGCUCACGCGCGCAGCCUCUUU-UCAGAUAGGGUU ---------......(((((....(-(((((((((((.(((....))).)))))))))))-)..))))).... ( -31.20) >psp.2242 4393819 63 - 5928787 ---------UUUCAACUGUCGCCCC-AAAGAGGCUGCCCGCUCAUGCGUGCGGCCUCUUUGUCGAAAGAGAAA ---------.(((...........(-(((((((((((.(((....))).))))))))))))((....))))). ( -30.30) >consensus _________UUUCAACUGUCGCCCC_AAAGAGGCUGCCCGCUCACGCGCGCAGCCUCUUUGUCAAAAGGGGAA ..........................(((((((((((.(((....))).)))))))))))............. (-24.79 = -24.18 + -0.61)

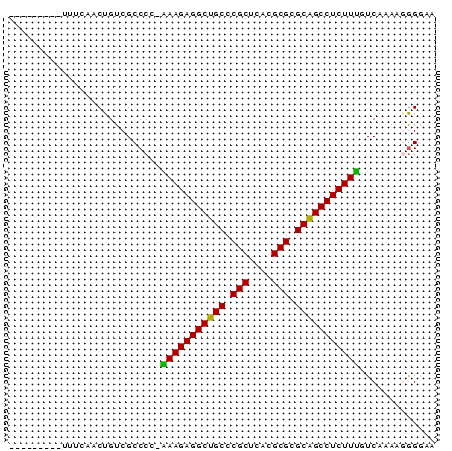
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:12:36 2007