
| Sequence ID | pao.2623 |
|---|---|
| Location | 4,956,424 – 4,956,733 |
| Length | 309 |
| Max. P | 0.953362 |

| Location | 4,956,424 – 4,956,543 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -44.09 |
| Consensus MFE | -38.99 |
| Energy contribution | -37.85 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
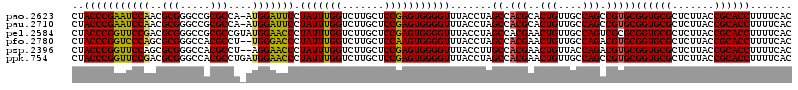
>pao.2623 4956424 119 + 6264404 CUACCCGAAUCCAACGCGGGCCGCGCCA-AUGGAUUCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGCACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCAC ......(((((((..(((.....)))..-.)))))))((((((((.......))))))))((((.....))))..((((.((.....)).))))((((((.......))))))....... ( -40.00) >pau.2710 5119982 119 + 6537648 CUACCCGAAUCCAACGCGGGCCGCGCCA-AUGGAUUCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGCACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCAC ......(((((((..(((.....)))..-.)))))))((((((((.......))))))))((((.....))))..((((.((.....)).))))((((((.......))))))....... ( -40.00) >pel.2584 4810264 120 + 5888780 CUACCCGGUUCCGACGCGGGCCGCGCCGUAUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGUCGCGCGGUGCGCUCUUACCGCACCUUUUCAC .....((((...((.(((.(((((((......(((((((((((((.......)))))))))))))..........((.(((....)))))))))))).)))))..))))........... ( -48.40) >pfo.2780 5286600 118 + 6438405 CUACCCGGUCCCAGCGCGGGCCACGCCU--UGGGACCCUAUUUGGUCUUGCUCCAAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCAC ......((((((((.(((.....))).)--)))))))((((((((.......))))))))((((.....))))..((((((....)))..((((((((......))))))))...))).. ( -46.30) >psp.2396 4704824 118 + 5928787 CUACCCGGUUCCAGCGCGGGCCACGCCU--AGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUUGCCACGAACUGUUACCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCAC ......(((..(((..(((((.......--..(((((((((((((.......))))))))))))).....))).))..)))..)))....((((((((......))))))))........ ( -46.14) >ppk.754 4668877 120 - 6181863 CUACCCGGUUCCGACGCGGGCCACGCCUGAUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCAC ......((((.((((.(((((...)))))...(((((((((((((.......)))))))))))))...............))))..))))((((((((......))))))))........ ( -43.70) >consensus CUACCCGGUUCCAACGCGGGCCACGCCU_AUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCAC ..(((((((((((..(((.....)))....))))))).(((((((.......))))))))))).......((.(((..(((....))).)))))((((((.......))))))....... (-38.99 = -37.85 + -1.14)

| Location | 4,956,424 – 4,956,543 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -44.85 |
| Consensus MFE | -44.29 |
| Energy contribution | -42.55 |
| Covariance contribution | -1.74 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
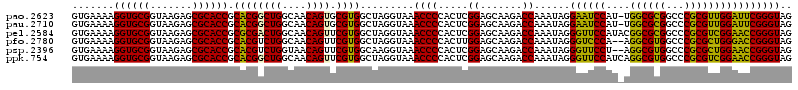
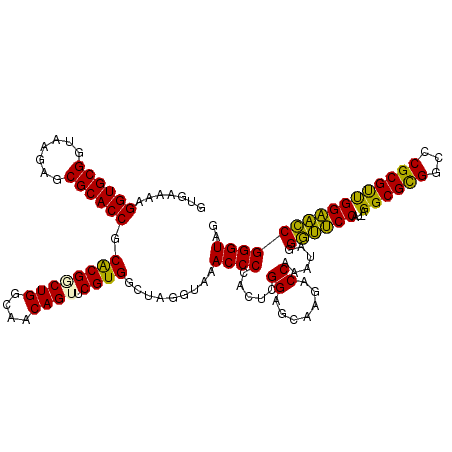
>pao.2623 4956424 119 - 6264404 GUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUGCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGAAUCCAU-UGGCGCGGCCCGCGUUGGAUUCGGGUAG .......((((((.......)))))).((((((((....)))).)))).........((((.....((.......))......(((((((.-..(((((...)))))))))))))))).. ( -43.70) >pau.2710 5119982 119 - 6537648 GUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUGCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGAAUCCAU-UGGCGCGGCCCGCGUUGGAUUCGGGUAG .......((((((.......)))))).((((((((....)))).)))).........((((.....((.......))......(((((((.-..(((((...)))))))))))))))).. ( -43.70) >pel.2584 4810264 120 - 5888780 GUGAAAAGGUGCGGUAAGAGCGCACCGCGCGACUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAUACGGCGCGGCCCGCGUCGGAACCGGGUAG .......((((((.......)))))).((((((((....))).)))))....((......))((((((..(..((((......)))).....(((((((...))))))))..)))))).. ( -46.10) >pfo.2780 5286600 118 - 6438405 GUGAAAAGGUGCGGUAAGAGCGCACCGCACGUCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUUGGAGCAAGACCAAAUAGGGUCCCA--AGGCGUGGCCCGCGCUGGGACCGGGUAG .......((((((.......)))))).((((.(((....)))..)))).........((((...((((.......))))....(((((((--..(((((...)))))))))))))))).. ( -48.00) >psp.2396 4704824 118 - 5928787 GUGAAAAGGUGCGGUAAGAGCGCACCGCACGUCUGGUAACAGUUCGUGGCAAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCU--AGGCGUGGCCCGCGCUGGAACCGGGUAG (((....((((((.......)))))).))).((((((..((((.((.(((..((......)).((.(((((....((.....))))))).--)).....))))).))))..))))))... ( -43.80) >ppk.754 4668877 120 + 6181863 GUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAUCAGGCGUGGCCCGCGUCGGAACCGGGUAG .......((((((.......)))))).((((((((....))).))))).........((((.....((.......))......((((((....((((((...)))))))))))))))).. ( -43.80) >consensus GUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAU_AGGCGCGGCCCGCGUUGGAACCGGGUAG .......((((((.......)))))).((((((((....)))).)))).........((((.....((.......))......((((((....((((((...)))))))))))))))).. (-44.29 = -42.55 + -1.74)
| Location | 4,956,543 – 4,956,662 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -37.08 |
| Energy contribution | -36.58 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2623 4956543 119 + 6264404 CCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCACCGCGCU- .......(((((....)))))...(((((.....((((.(((....(((((((....(((((.....))))).....(((.....))))))))))))))))).........)))))...- ( -42.54) >pau.2710 5120101 119 + 6537648 CCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCACCGCGCU- .......(((((....)))))...(((((.....((((.(((....(((((((....(((((.....))))).....(((.....))))))))))))))))).........)))))...- ( -42.54) >pel.2584 4810384 118 + 5888780 CCUUACCGGCGCCGAAGCGCUUAGGCGGUUGUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCCCC-GCCU- .......(((((....))))).((((((......((((.(((....(((((((....(((((.....))))).....(((.....)))))))))))))))))..........))-))))- ( -43.29) >pf5.3093 5834063 118 + 7074893 CCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCCCCCAGGCAUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCCCCC-CUCU- .....(((((....(((((((...((((.......)))))))))))((((((((....((....(((((.....))))).....)).))))))))).)))).............-....- ( -35.90) >pfo.2780 5286718 118 + 6438405 CCUUACCGGCGCCGAAGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCUCCCAGGCAUUACCUGGCACUUCGCCCUAUGGAGCCCGGACUUUCCUCCCCC-CCCU- .......(((((....)))))..((.((......((((.(((....((((((((....((....(((((.....))))).....)).)))))))))))))))....)).))...-....- ( -36.40) >ppk.754 4668997 119 - 6181863 CCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCUCCCAGGCGUUACCUGGCACUCCGCCCUGUGGAGCCCGGACUUUCCUCCCCC-UGCUG ......(((((...(((((((...((((.......)))))))))))((((..((((((((((.....)))))(((..(((.....)))..))))))))))))............-))))) ( -41.00) >consensus CCUUACCGGCGCCGAAGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCUCCCAGGCGUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCCCCC_CGCU_ .......(((((....)))))..((.((......((((.(((....(((((((....(((((.....))))).....(((.....)))))))))))))))))....)).))......... (-37.08 = -36.58 + -0.50)
| Location | 4,956,543 – 4,956,662 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.88 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -42.23 |
| Energy contribution | -41.65 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2623 4956543 119 - 6264404 -AGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGG -((.(((((.(.((....((((.((((....))))......(((((.....))))).((((....)))).)))).....)).)........)))))))..((((....))))........ ( -43.20) >pau.2710 5120101 119 - 6537648 -AGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGG -((.(((((.(.((....((((.((((....))))......(((((.....))))).((((....)))).)))).....)).)........)))))))..((((....))))........ ( -43.20) >pel.2584 4810384 118 - 5888780 -AGGC-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAACAACCGCCUAAGCGCUUCGGCGCCGGUAAGG -((((-((..(.......(((((((...((..(((.....)))..))...))))))).(((((...((...))...)))))........)..))))))..((((....))))........ ( -50.10) >pf5.3093 5834063 118 - 7074893 -AGAG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGGAGUGCCAGGUAAUGCCUGGGGGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGG -....-.((((.((.....))...))))...((((((.((.(((((.....))))).((((....))))))((......))...........))))))..((((....))))........ ( -46.80) >pfo.2780 5286718 118 - 6438405 -AGGG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGAAGUGCCAGGUAAUGCCUGGGAGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCUUCGGCGCCGGUAAGG -((((-(((((.((.....))...))))....((((.....(((((.....))))).((((....))))........))))...........)).)))..((((....))))........ ( -46.50) >ppk.754 4668997 119 + 6181863 CAGCA-GGGGGAGGAAAGUCCGGGCUCCACAGGGCGGAGUGCCAGGUAACGCCUGGGAGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGG .....-.((((.((.....))...))))...((((((.((.(((((.....))))).((((....))))))((......))...........))))))..((((....))))........ ( -46.80) >consensus _AGCG_GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCUUCGGCGCCGGUAAGG ............((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))..........((((.....((((....)))))))).... (-42.23 = -41.65 + -0.58)

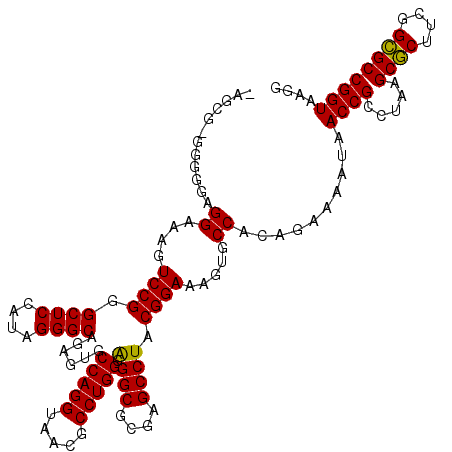

| Location | 4,956,583 – 4,956,677 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -23.07 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.855270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2623 4956583 94 + 6264404 GCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCACCGCGCU----------------AUUGC----------GCGACAGCGA ((....((((..((((((((((.....))))).....(((.....))).....)))))))))...........(((((.----------------...))----------)))...)).. ( -36.90) >pau.2710 5120141 94 + 6537648 GCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCACCGCGCU----------------AUUGC----------GCGACAGCGA ((....((((..((((((((((.....))))).....(((.....))).....)))))))))...........(((((.----------------...))----------)))...)).. ( -36.90) >pel.2584 4810424 103 + 5888780 GCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCCCCC-GCCU----------------CUUGCGGAACAAAAGACGGCAGCGA ((....(((((((....(((((.....))))).....(((.....))))))))))(((.(((......))).((-((..----------------...))))..........))).)).. ( -35.50) >pf5.3093 5834103 99 + 7074893 GCACUUUCCGUAGGCUCACGCCCCCCAGGCAUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCCCCC-CUCU----------------AAUUU----UCAUAGAGGGCAGCGA ((....((((..(((((..(((.....))).......(((.....))).....)))))))))..........((-((((----------------(....----...)))))))..)).. ( -33.30) >pfo.2780 5286758 99 + 6438405 GCACUUUCCGUAGGCUCACGCCUCCCAGGCAUUACCUGGCACUUCGCCCUAUGGAGCCCGGACUUUCCUCCCCC-CCCU----------------AAUUU----UCAUAGAGGGCAGCGA ((....((((((((....((....(((((.....))))).....)).))))))))(((((((......)))...-..((----------------(....----...))).)))).)).. ( -30.60) >ppk.754 4669037 119 - 6181863 GCACUUUCCGUAGGCUCACGCCUCCCAGGCGUUACCUGGCACUCCGCCCUGUGGAGCCCGGACUUUCCUCCCCC-UGCUGCUCACACAACAAAAAGUUGCGGAACAAAAACAGGCAGCGA ......((((..((((((((((.....)))))(((..(((.....)))..))))))))))))............-(((((((..(.((((.....)))).)...........))))))). ( -37.02) >consensus GCACUUUCCGUAGGCUCACGCCUCCCAGGCGUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCCCCC_CGCU________________AUUGC______A_AGACGGCAGCGA ......((((..((((((((((.....))))).....(((.....))).....))))))))).......................................................... (-23.07 = -23.15 + 0.08)

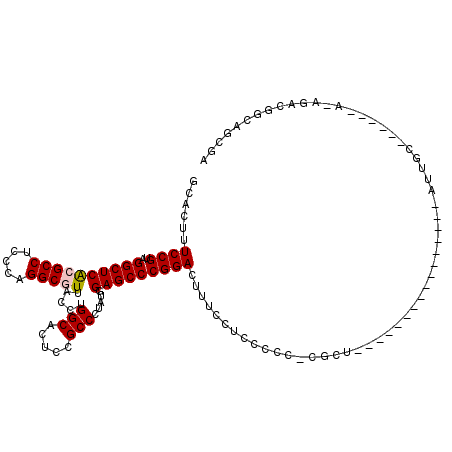
| Location | 4,956,583 – 4,956,677 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -44.32 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.08 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2623 4956583 94 - 6264404 UCGCUGUCGC----------GCAAU----------------AGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGC ((.(..((((----------((...----------------.))))))..).))....((((.((((....))))......(((((.....))))).((((....)))).))))...... ( -40.90) >pau.2710 5120141 94 - 6537648 UCGCUGUCGC----------GCAAU----------------AGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGC ((.(..((((----------((...----------------.))))))..).))....((((.((((....))))......(((((.....))))).((((....)))).))))...... ( -40.90) >pel.2584 4810424 103 - 5888780 UCGCUGCCGUCUUUUGUUCCGCAAG----------------AGGC-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGC ((.((.((((((((((.....))))----------------))))-)).)).))....((((.((((....))))......(((((.....))))).((((....)))).))))...... ( -45.20) >pf5.3093 5834103 99 - 7074893 UCGCUGCCCUCUAUGA----AAAUU----------------AGAG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGGAGUGCCAGGUAAUGCCUGGGGGGCGUGAGCCUACGGAAAGUGC ((.((.(((((((...----....)----------------))))-)).)).))....((((.(((((.......))))).(((((.....))))).((((....)))).))))...... ( -44.50) >pfo.2780 5286758 99 - 6438405 UCGCUGCCCUCUAUGA----AAAUU----------------AGGG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGAAGUGCCAGGUAAUGCCUGGGAGGCGUGAGCCUACGGAAAGUGC ((.((.(((((((...----....)----------------))))-)).)).))....((((.((((....))))......(((((.....))))).((((....)))).))))...... ( -41.20) >ppk.754 4669037 119 + 6181863 UCGCUGCCUGUUUUUGUUCCGCAACUUUUUGUUGUGUGAGCAGCA-GGGGGAGGAAAGUCCGGGCUCCACAGGGCGGAGUGCCAGGUAACGCCUGGGAGGCGUGAGCCUACGGAAAGUGC ((.((.(((((...(((((((((((.....)))))).))))))))-)).)).))....((((.(((((.......))))).(((((.....))))).((((....)))).))))...... ( -53.20) >consensus UCGCUGCCGUCU_U______GCAAU________________AGCG_GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGC .((((.(((((...................................))))).......((((.((((....))))......(((((.....))))).((((....)))).)))).)))). (-30.42 = -30.08 + -0.33)
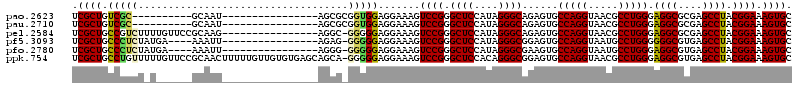
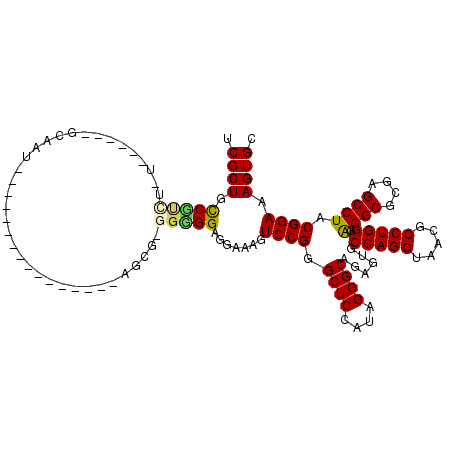
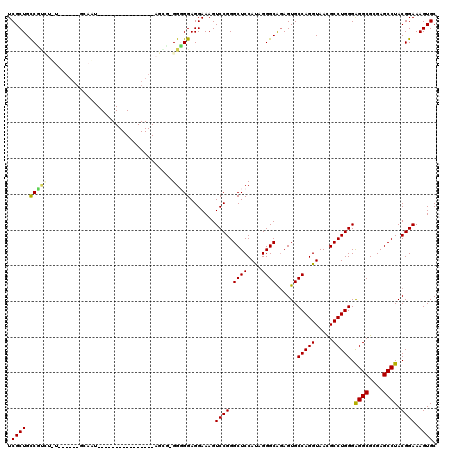
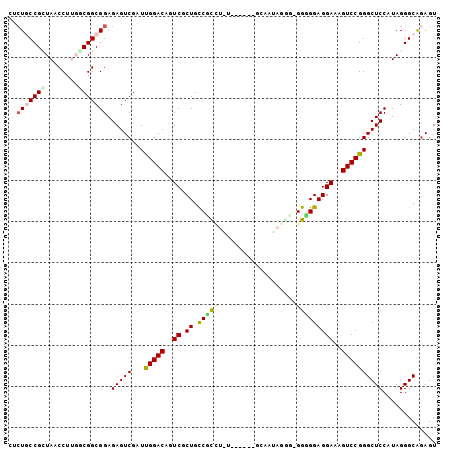
| Location | 4,956,623 – 4,956,717 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.36 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -25.93 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.2623 4956623 94 - 6264404 UGCUCCCGCUAACCUUGUCGGCGGAGAGUCGAUUGGACAGUCGCUGUCGC----------GCAAUAGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGU (((((((((..((...))..)))).(((((...(((((..((.(..((((----------((....))))))..)..))..))))))))))....))))).... ( -41.90) >pau.2710 5120181 94 - 6537648 UGCUCCCGCUAACCUUGUCGGCGGAGAGUCGAUUGGACAGUCGCUGUCGC----------GCAAUAGCGCGGUGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGU (((((((((..((...))..)))).(((((...(((((..((.(..((((----------((....))))))..)..))..))))))))))....))))).... ( -41.90) >pel.2584 4810464 103 - 5888780 GCCUGCCGGUAACCUUGUCGGCGGAGAGUCGAUCGGACAGUCGCUGCCGUCUUUUGUUCCGCAAGAGGC-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGU ((((((((..(....)..))))((((......((((((..((.((.((((((((((.....))))))))-)).))..))..)))))).))))...))))..... ( -49.70) >pf5.3093 5834143 99 - 7074893 CUCUGCCGUUAACCUGCGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCCUCUAUGA----AAAUUAGAG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGGAGU (((((((((........)))))))))......((((((..((.((.(((((((...----....)))))-)).))..))..))))))(((((.......))))) ( -45.00) >pfo.2780 5286798 99 - 6438405 CUCUGCCGUUAACCUGCGCGGCGGAGAGUCGAUCGGACAGUCGCUGCCCUCUAUGA----AAAUUAGGG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGAAGU (((((((((........)))))))))..((..((((((..((.((.(((((((...----....)))))-)).))..))..))))))((((....))))))... ( -44.70) >psp.2396 4705022 103 - 5928787 AUGUGCCGCUAACCUUGGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCUCUUGUUGUUCCGCAACAGGG-GGGGGAGGAAAGUCCGGGCUCCAUAGGGCGGAGU .((((((((((....)))))))((((......((((((..((.((.((((((((((.....))))))))-)).))..))..)))))).)))).....))).... ( -46.30) >consensus CUCUGCCGCUAACCUUGGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCGCCU_U______GCAAUAGGG_GGGGGAGGAAAGUCCGGGCUCCAUAGGGCAGAGU ..(((((((........))))))).(((((...(((((..((.((.(((((...................)))))))))..))))))))))............. (-25.93 = -25.88 + -0.05)
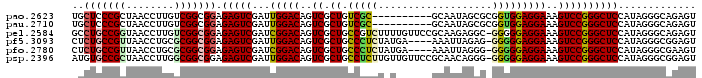
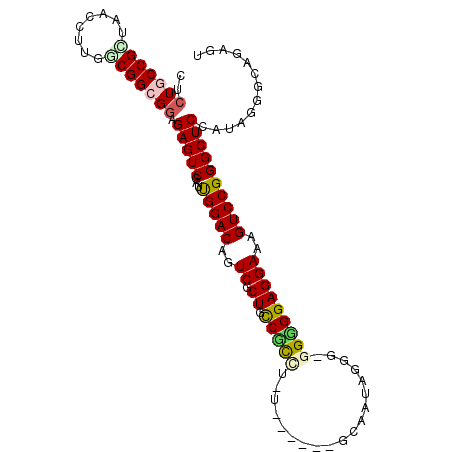
| Location | 4,956,639 – 4,956,733 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -24.84 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
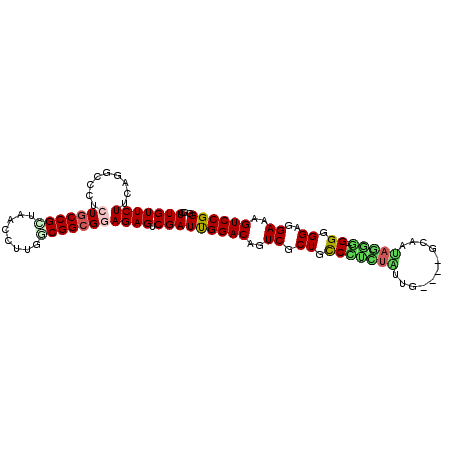
>pao.2623 4956639 94 - 6264404 GCUUGUUCUUCGCGCCUGCUCCCGCUAACCUUGUCGGCGGAGAGUCGAUUGGACAGUCGCUGUCGC----------GCAAUAGCGCGGUGGAGGAAAGUCCGGG ((.........)).((.((((((((..((...))..)))).)))).....((((..((.(..((((----------((....))))))..)..))..)))))). ( -37.30) >pel.2584 4810480 103 - 5888780 GCUUGUUCUUGAGCGCGCCUGCCGGUAACCUUGUCGGCGGAGAGUCGAUCGGACAGUCGCUGCCGUCUUUUGUUCCGCAAGAGGC-GGGGGAGGAAAGUCCGGG ((((......))))((.(((((((..(....)..)))))).).))...((((((..((.((.((((((((((.....))))))))-)).))..))..)))))). ( -44.90) >pf5.3093 5834159 99 - 7074893 GCUUGUUCUUCGUUCGCUCUGCCGUUAACCUGCGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCCUCUAUGA----AAAUUAGAG-GGGGGAGGAAAGUCCGGG .........(((....(((((((((........)))))))))...)))((((((..((.((.(((((((...----....)))))-)).))..))..)))))). ( -42.30) >pfo.2780 5286814 99 - 6438405 GCUUGUUCUUCGGCCGCUCUGCCGUUAACCUGCGCGGCGGAGAGUCGAUCGGACAGUCGCUGCCCUCUAUGA----AAAUUAGGG-GGGGGAGGAAAGUCCGGG .........(((((..(((((((((........))))))))).)))))((((((..((.((.(((((((...----....)))))-)).))..))..)))))). ( -46.70) >psp.2396 4705038 103 - 5928787 GCUUGUUCUUGAAGGCAUGUGCCGCUAACCUUGGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCUCUUGUUGUUCCGCAACAGGG-GGGGGAGGAAAGUCCGGG ((((........)))).(.((((((((....)))))))).).......((((((..((.((.((((((((((.....))))))))-)).))..))..)))))). ( -44.10) >pst.2629 4986698 103 - 6397126 GCUUGUUCUUGAUGGCGUGUGCCGCUAACCUUGUCGGCGGAGAGUCGAUUGGACAGUCGCUGCCUUUUGUUGUUCCGCAACAAGG-GGGGGAGGAAAGUCCGGG ............(((((.....))))).((..((((((.....)))))).((((..((.((.((((((((((.....))))))))-)).))..))..)))))). ( -38.30) >consensus GCUUGUUCUUCAGGCCCUCUGCCGCUAACCUUGGCGGCGGAGAGUCGAUUGGACAGUCGCUGCCCUCUAUUG____GCAAUAGGG_GGGGGAGGAAAGUCCGGG ..(((((((.........(((((((........))))))))))).)))((((((..((.((.(((((((...........))))).)).))..))..)))))). (-24.84 = -24.65 + -0.19)

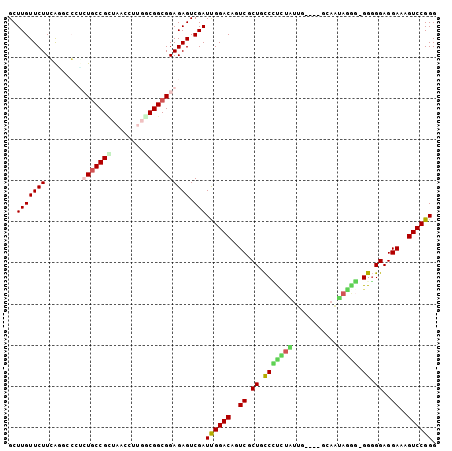
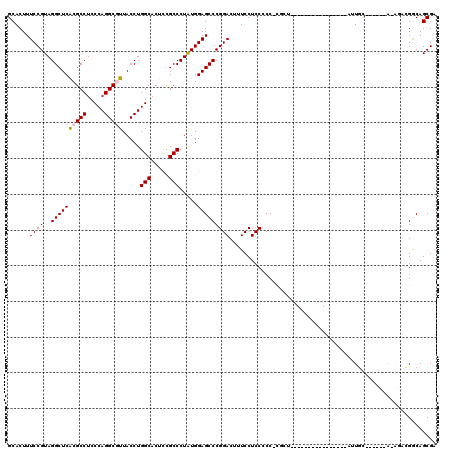
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:12:27 2007