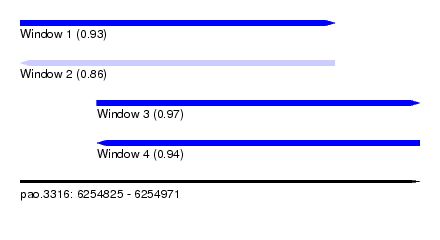
| Sequence ID | pao.3316 |
|---|---|
| Location | 6,254,825 – 6,254,971 |
| Length | 146 |
| Max. P | 0.965369 |

| Location | 6,254,825 – 6,254,940 |
|---|---|
| Length | 115 |
| pre Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.05 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3316 6254825 115 + 6264404 UCUCAG-UCUUCUGGUGUCGGCACUGCC--A-AA-UCAACGACUUGGCAUAAAAAGUGCCCACAAAAUAGCGCGCAGAGUAUAGGGGGGGAGUCCCCCCUGUUCAACUGGCUGGUAGUGA ..((((-((.((((((((.(((((((((--(-(.-((...)).)))))......)))))).........)))).))))(.(((((((((....))))))))).)....))))))...... ( -43.60) >pf5.3746 7063014 112 + 7074893 ---UUGCUCCUCAAGGGUCGG--CUGCC--AGAAUUCAUUAACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGCGGUUCAGCCCCUAUUCAACUGCCGGGUAGUGA ---.((..((....))..))(--(((((--......((((...(((((((.....)))))))...)))-)((.((((...((((((((......))))))))....)))))))))))).. ( -42.80) >pst.3346 6388004 111 + 6397126 ---CUGCUCCUCUGAUGUCGG--UCGCC--AUAA-UCAAUAACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGCGGUUCAGCCCCUAUUCAACAGGCGGGUAGUGA ---((((((.((((.((((((--(.(((--(...-.........))))((.....)))))))))....-.........(.((((((((......)))))))).)..)))).))))))... ( -39.50) >pfo.3375 6429539 112 + 6438405 ---UUGCUCUCCAAUGGUCGG--CUGCC--GAAAUUCAAUAACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGCGGUUCUGCCCCUAUUCAACUGUCAGGUAGUGA ---(..((..((....(((((--(((((--((...........))))))........)))))).....-....((((...(((((((((....)))))))))....))))..)).))..) ( -38.20) >ppk.0 2 113 + 6181863 ---CUGCUCCUCGGAAGUCGA--CCAACAAGUCA-GCUAUGACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGUGGAUUAACCCCUAUUCAACUCUUCGGUAGUGA ---((((.........((((.--....(((((((-....)))))))((((.....)))))))).....-...((.(((((((((((((......))))))))...))))).))))))... ( -40.20) >pel.3210 5879873 113 + 5888780 ---CUGCUCCUCGAAAGUCGA--CCGCCUAGUCA-GCAAUGACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGUGGAUUAACCCCUAUUCAACUCUUCGGUAGUGA ---((((((..(....)..))--.(((...((((-....))))(((((((.....)))))))......-)))((.(((((((((((((......))))))))...))))).))))))... ( -40.30) >consensus ___CUGCUCCUCAAAAGUCGG__CCGCC__AUAA_UCAAUAACUUGGCAUAAUUUGUGCCGACAAAAU_GCGCGCAGAGUAUAGGGGCGGAUCAGCCCCUAUUCAACUGUCCGGUAGUGA ...((((((..(....)..))......................(((((((.....)))))))...........)))).(.((((((((......)))))))).)................ (-24.74 = -24.05 + -0.69)
| Location | 6,254,825 – 6,254,940 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -37.69 |
| Consensus MFE | -23.38 |
| Energy contribution | -22.88 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3316 6254825 115 - 6264404 UCACUACCAGCCAGUUGAACAGGGGGGACUCCCCCCCUAUACUCUGCGCGCUAUUUUGUGGGCACUUUUUAUGCCAAGUCGUUGA-UU-U--GGCAGUGCCGACACCAGAAGA-CUGAGA .........(((((......(((((((....))))))).....))).)).......(((.((((((......((((((((...))-))-)--))))))))).))).(((....-)))... ( -43.60) >pf5.3746 7063014 112 - 7074893 UCACUACCCGGCAGUUGAAUAGGGGCUGAACCGCCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUUAAUGAAUUCU--GGCAG--CCGACCCUUGAGGAGCAA--- ..........((((..(.((((((((......)))))))).).))))..((-.....((((((........(((((((((....)))).)--)))))--)))))((....)).))..--- ( -39.50) >pst.3346 6388004 111 - 6397126 UCACUACCCGCCUGUUGAAUAGGGGCUGAACCGCCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUUAUUGA-UUAU--GGCGA--CCGACAUCAGAGGAGCAG--- ...........(((((..((((((((......)))))))).(((((.....-....((((((((.......))))..(((((...-..))--)))..--..)))).))))).)))))--- ( -34.20) >pfo.3375 6429539 112 - 6438405 UCACUACCUGACAGUUGAAUAGGGGCAGAACCGCCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUUAUUGAAUUUC--GGCAG--CCGACCAUUGGAGAGCAA--- .........(.(((..(.((((((((......)))))))).).))))((.(-.....((((((........(((((((((....))))).--)))))--))))).....)...))..--- ( -35.00) >ppk.0 2 113 - 6181863 UCACUACCGAAGAGUUGAAUAGGGGUUAAUCCACCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUCAUAGC-UGACUUGUUGG--UCGACUUCCGAGGAGCAG--- .......((.(((((...((((((((......))))))))))))).)).((-.....((((((.(((.......(((((((....-)))))))))))--))))).((....))))..--- ( -36.91) >pel.3210 5879873 113 - 5888780 UCACUACCGAAGAGUUGAAUAGGGGUUAAUCCACCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUCAUUGC-UGACUAGGCGG--UCGACUUUCGAGGAGCAG--- .......((.(((((...((((((((......))))))))))))).)).((-.(((((..((((.......))))((((((((((-(.....)))))--).))))).))))).))..--- ( -36.90) >consensus UCACUACCCGACAGUUGAAUAGGGGCUAAACCGCCCCUAUACUCUGCGCGC_AUUUUGUCGGCACAAAUUAUGCCAAGUCAUUGA_UUAC__GGCAG__CCGACAUCAGAGGAGCAG___ .............((((.((((((((......))))))))...(((((((......))).((((.......))))..................))))...))))................ (-23.38 = -22.88 + -0.50)

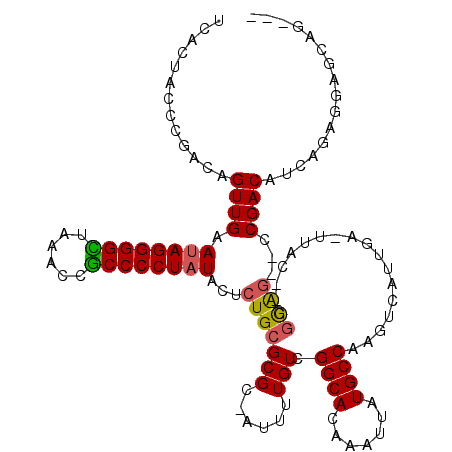
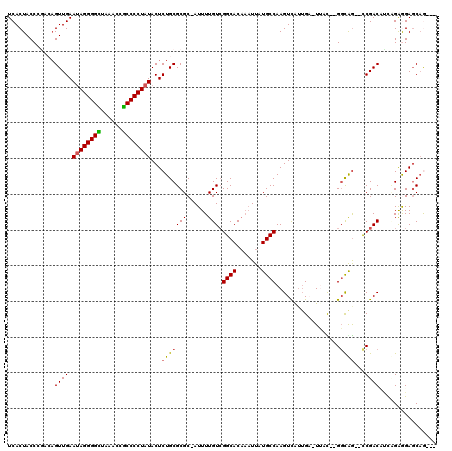
| Location | 6,254,853 – 6,254,971 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -29.64 |
| Energy contribution | -29.42 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3316 6254853 118 + 6264404 -AA-UCAACGACUUGGCAUAAAAAGUGCCCACAAAAUAGCGCGCAGAGUAUAGGGGGGGAGUCCCCCCUGUUCAACUGGCUGGUAGUGAUUUCCGACUACCCGCUACAACCCAAAGUGUG -..-...((.(((((((((.....))))).......((((((.(((...(((((((((....)))))))))....))))).((((((........)))))).)))).......)))))). ( -42.00) >pau.3455 6528109 118 + 6537648 -AA-UCAACGACUUGGCAUAAAAAGUGCCCACAAAAUAGCGCGCAGAGUAUAGGGGGGGAGUCCCCCCUGUUCAACUGGCUGGUAGUGAUUUCCGACUACCCGCUACAACCCAAAGUGUG -..-...((.(((((((((.....))))).......((((((.(((...(((((((((....)))))))))....))))).((((((........)))))).)))).......)))))). ( -42.00) >pf5.3746 7063038 107 + 7074893 GAAUUCAUUAACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGCGGUUCAGCCCCUAUUCAACUGCCGGGUAGUGAUUUCCGACUGCGCGCUACA------------ ............(((((((.....)))))))......-((((((((.(.((((((((......)))))))).)......((((........)))).))))))))....------------ ( -44.80) >pfo.3375 6429563 107 + 6438405 AAAUUCAAUAACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGCGGUUCUGCCCCUAUUCAACUGUCAGGUAGUGAUUUCCGACUGCGCGCUACA------------ ............(((((((.....)))))))......-((((((((.(.(((((((((....))))))))).).((((.....)))).........))))))))....------------ ( -41.40) >ppk.0 28 106 + 6181863 UCA-GCUAUGACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGUGGAUUAACCCCUAUUCAACUCUUCGGUAGUGAUUUCCGACUUCACGCUACA------------ ...-((......(((((((.....)))))))......-)).((.(((((((((((((......))))))))...))))).))((((((.............)))))).------------ ( -35.92) >pel.3210 5879899 106 + 5888780 UCA-GCAAUGACUUGGCAUAAUUUGUGCCGACAAAAU-GCGCGCAGAGUAUAGGGGUGGAUUAACCCCUAUUCAACUCUUCGGUAGUGAUUUCCGACUGCGCGCUACA------------ (((-....))).(((((((.....)))))))......-((((((((.(.((((((((......)))))))).)......((((.........))))))))))))....------------ ( -43.60) >consensus _AA_UCAAUGACUUGGCAUAAUUUGUGCCGACAAAAU_GCGCGCAGAGUAUAGGGGCGGAUUAACCCCUAUUCAACUGUCCGGUAGUGAUUUCCGACUGCGCGCUACA____________ ............(((((((.....))))))).........((((((...((((((((......))))))))....)))....(((((........))))).)))................ (-29.64 = -29.42 + -0.22)
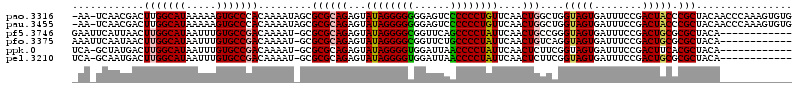
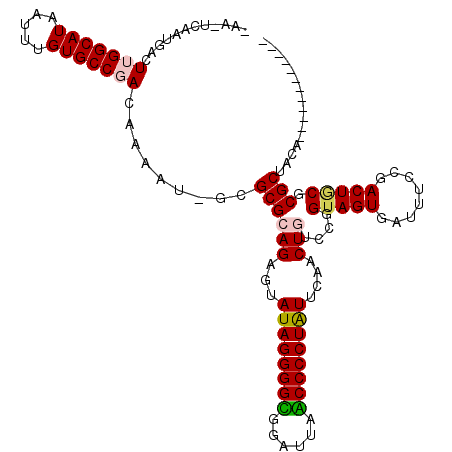
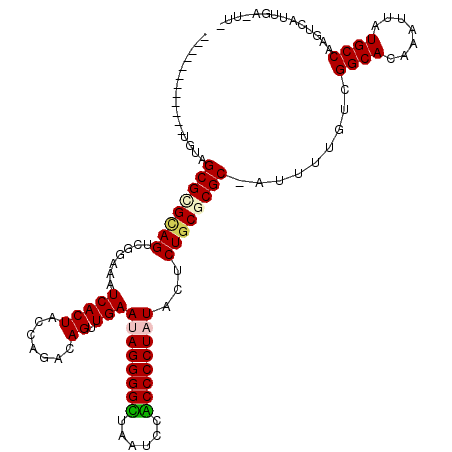
| Location | 6,254,853 – 6,254,971 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.87 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.22 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao.3316 6254853 118 - 6264404 CACACUUUGGGUUGUAGCGGGUAGUCGGAAAUCACUACCAGCCAGUUGAACAGGGGGGACUCCCCCCCUAUACUCUGCGCGCUAUUUUGUGGGCACUUUUUAUGCCAAGUCGUUGA-UU- ..(((........(((((.((((((........)))))).(((((......(((((((....))))))).....))).)))))))...)))((((.......))))..........-..- ( -39.70) >pau.3455 6528109 118 - 6537648 CACACUUUGGGUUGUAGCGGGUAGUCGGAAAUCACUACCAGCCAGUUGAACAGGGGGGACUCCCCCCCUAUACUCUGCGCGCUAUUUUGUGGGCACUUUUUAUGCCAAGUCGUUGA-UU- ..(((........(((((.((((((........)))))).(((((......(((((((....))))))).....))).)))))))...)))((((.......))))..........-..- ( -39.70) >pf5.3746 7063038 107 - 7074893 ------------UGUAGCGCGCAGUCGGAAAUCACUACCCGGCAGUUGAAUAGGGGCUGAACCGCCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUUAAUGAAUUC ------------....((((((((((((..........)))))......((((((((......))))))))....)))))))-........((((.......)))).............. ( -40.70) >pfo.3375 6429563 107 - 6438405 ------------UGUAGCGCGCAGUCGGAAAUCACUACCUGACAGUUGAAUAGGGGCAGAACCGCCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUUAUUGAAUUU ------------....((((((((((((..........)))))......((((((((......))))))))....)))))))-........((((.......)))).............. ( -39.20) >ppk.0 28 106 - 6181863 ------------UGUAGCGUGAAGUCGGAAAUCACUACCGAAGAGUUGAAUAGGGGUUAAUCCACCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUCAUAGC-UGA ------------....(((((...((((.........))))(((((...((((((((......))))))))))))).)))))-........((((.......))))..........-... ( -33.00) >pel.3210 5879899 106 - 5888780 ------------UGUAGCGCGCAGUCGGAAAUCACUACCGAAGAGUUGAAUAGGGGUUAAUCCACCCCUAUACUCUGCGCGC-AUUUUGUCGGCACAAAUUAUGCCAAGUCAUUGC-UGA ------------.(((((((((..((((.........))))(((((...((((((((......)))))))))))))))))))-........((((.......)))).......)))-... ( -39.30) >consensus ____________UGUAGCGCGCAGUCGGAAAUCACUACCAGACAGUUGAAUAGGGGCUAAUCCACCCCUAUACUCUGCGCGC_AUUUUGUCGGCACAAAUUAUGCCAAGUCAUUGA_UU_ ................((((((((.......(((((.......)).)))((((((((......))))))))...)))))))).........((((.......)))).............. (-30.30 = -30.22 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 21:03:17 2007