
| Sequence ID | pao_1 |
|---|---|
| Location | 5,537,740 – 5,537,794 |
| Length | 54 |
| Max. P | 0.999996 |

| Location | 5,537,740 – 5,537,794 |
|---|---|
| Length | 54 |
| Sequences | 6 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
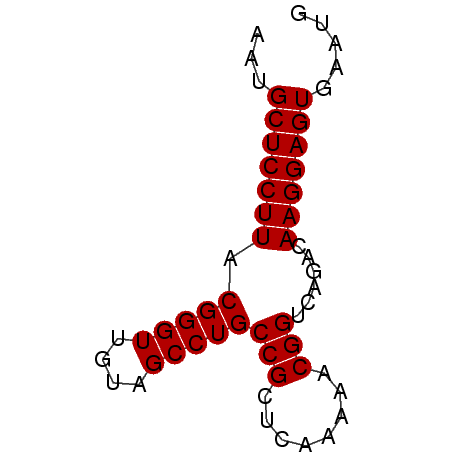
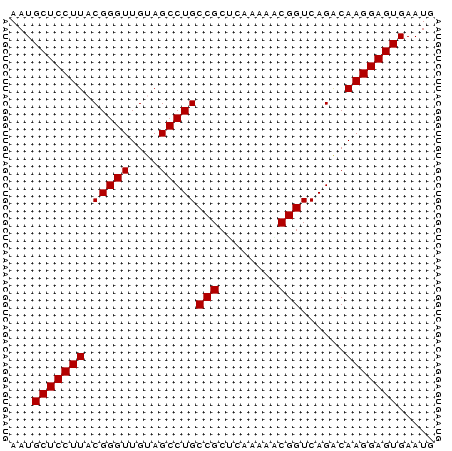
>pao_1 5537740 54 + 1 AAUGCUCCUUACGGGUUGCAGCCUGCCGCA---GAACGGUCAGGCAAGGAGUGAAUG ...(((((((..(.....).((((((((..---...))).))))))))))))..... ( -21.00) >pau_1 1 54 + 1 AAUGCUCCUUACGGGUUGCAGCCUGCCGCA---GAACGGUCAGGCAAGGAGUGAAUG ...(((((((..(.....).((((((((..---...))).))))))))))))..... ( -21.00) >pf5_1 1 57 + 1 AAUGCUCCUUACGGGUUGUAGCCUGCCGCCUAUAAGCGGUCAGACAAGGAGUGAAUG ...(((((((.(((((....)))))((((......))))......)))))))..... ( -20.30) >pfo_1 1 57 + 1 AAUGCUCCUUACGGGUUGUAGCCUGCCGCUCAAAAACGGUCAGACAAGGAGUGAAUG ...(((((((.(((((....)))))(((........)))......)))))))..... ( -16.80) >pfs_1 1 57 + 1 AAUGCUCCUUACGGGUUGUAGCCUGCCGCUCAAAAACGGUCAGACAAGGAGUGAAUG ...(((((((.(((((....)))))(((........)))......)))))))..... ( -16.80) >psp_1 1 57 + 1 AAUGCUCCUUACGGGUUGUAGCCUGCCGCUCAAAAGCGGUCAGACAAGGAGUGAAUG ...(((((((.(((((....)))))(((((....)))))......)))))))..... ( -21.40) >consensus AAUGCUCCUUACGGGUUGUAGCCUGCCGCUCAAAAACGGUCAGACAAGGAGUGAAUG ...(((((((.(((((....)))))(((........)))......)))))))..... (-17.23 = -17.23 + -0.00)

| Location | 5,537,740 – 5,537,794 |
|---|---|
| Length | 54 |
| Sequences | 6 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -20.05 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.03 |
| Structure conservation index | 1.04 |
| SVM decision value | 5.97 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
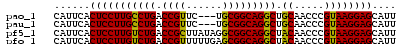
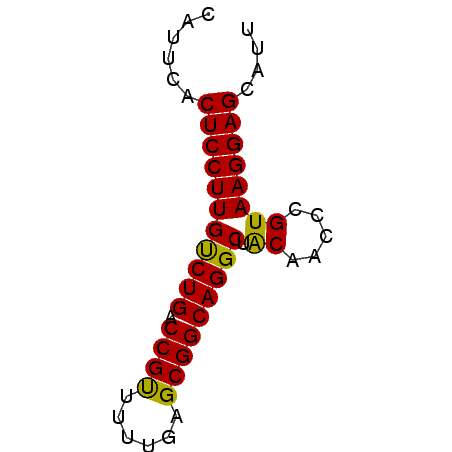
>pao_1 5537740 54 - 1 CAUUCACUCCUUGCCUGACCGUUC---UGCGGCAGGCUGCAACCCGUAAGGAGCAUU ......(((((((((((.(((...---..)))))))).((.....)))))))).... ( -20.30) >pau_1 1 54 - 1 CAUUCACUCCUUGCCUGACCGUUC---UGCGGCAGGCUGCAACCCGUAAGGAGCAUU ......(((((((((((.(((...---..)))))))).((.....)))))))).... ( -20.30) >pf5_1 1 57 - 1 CAUUCACUCCUUGUCUGACCGCUUAUAGGCGGCAGGCUACAACCCGUAAGGAGCAUU ......(((((((((((.(((((....)))))))))).((.....)))))))).... ( -21.20) >pfo_1 1 57 - 1 CAUUCACUCCUUGUCUGACCGUUUUUGAGCGGCAGGCUACAACCCGUAAGGAGCAUU ......(((((((((((.(((((....)))))))))).((.....)))))))).... ( -18.70) >pfs_1 1 57 - 1 CAUUCACUCCUUGUCUGACCGUUUUUGAGCGGCAGGCUACAACCCGUAAGGAGCAUU ......(((((((((((.(((((....)))))))))).((.....)))))))).... ( -18.70) >psp_1 1 57 - 1 CAUUCACUCCUUGUCUGACCGCUUUUGAGCGGCAGGCUACAACCCGUAAGGAGCAUU ......(((((((((((.(((((....)))))))))).((.....)))))))).... ( -21.10) >consensus CAUUCACUCCUUGUCUGACCGUUUUUGAGCGGCAGGCUACAACCCGUAAGGAGCAUU ......(((((((((((.((((......))))))))).((.....)))))))).... (-20.82 = -20.15 + -0.67)
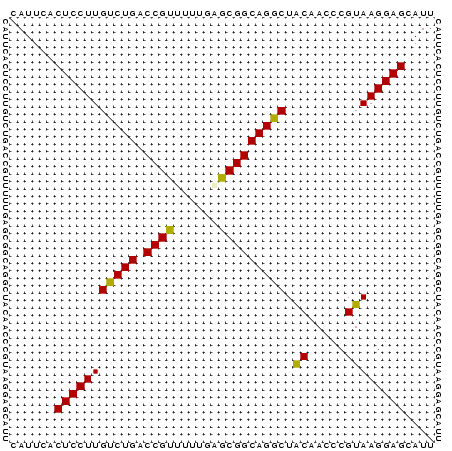
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:53:31 2007