
| Sequence ID | pao_1 |
|---|---|
| Location | 5,284,038 – 5,284,104 |
| Length | 66 |
| Max. P | 0.959949 |

| Location | 5,284,038 – 5,284,104 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | forward |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -15.51 |
| Energy contribution | -16.72 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
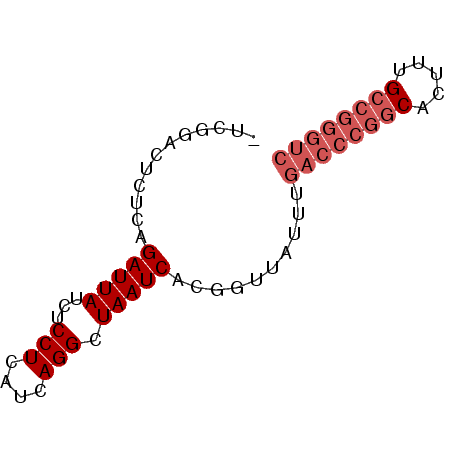
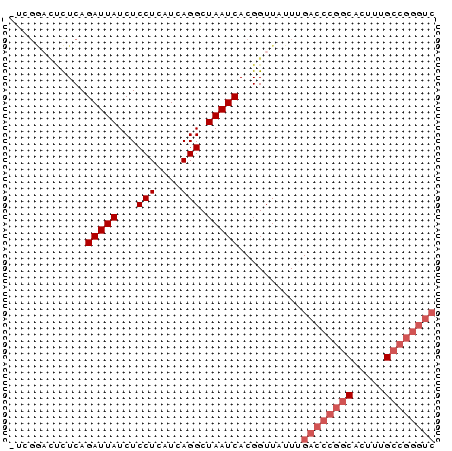
>pao_1 5284038 66 + 1 UCGGACUCUUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU-UUUGACCCGGCACUUUGCCGGGUC ............(((((...(((....))).)))))......-...((((((((.....)))))))) ( -20.60) >pel_1 1 65 + 1 -UCGGACUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUU-GACCCGGCAACUUGCCGGGUC -...((((....(((((...(((....))).)))))..))))...-((((((((.....)))))))) ( -21.20) >pf5_1 1 65 + 1 -UCGGACUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUUUGACCCGGCUCUU-GCCGGGUC -...((((....(((((...(((....))).)))))..))))....((((((((....-)))))))) ( -21.40) >pfo_1 1 52 + 1 -UCGGACUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUUUGACCCGG-------------- -.((........(((((...(((....))).)))))..((((....)))))).-------------- ( -7.70) >ppk_1 1 65 + 1 -UCGGACUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUU-GACCCGGCAAUUUGCCGGGUC -...((((....(((((...(((....))).)))))..))))...-((((((((.....)))))))) ( -21.20) >pst_1 1 58 + 1 --------UUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUU-UUUGACCCGGCUCUUUGCCGGGUC --------....(((((...(((....))).)))))......-...((((((((.....)))))))) ( -20.80) >consensus _UCGGACUCUCAGAUUAUCUCCUCAUCAGGCUAAUCACGGUUAUUUGACCCGGCACUUUGCCGGGUC ............(((((...(((....))).)))))..........((((((((.....)))))))) (-15.51 = -16.72 + 1.21)

| Location | 5,284,038 – 5,284,104 |
|---|---|
| Length | 66 |
| Sequences | 6 |
| Columns | 67 |
| Reading direction | reverse |
| Mean pairwise identity | 82.16 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.43 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
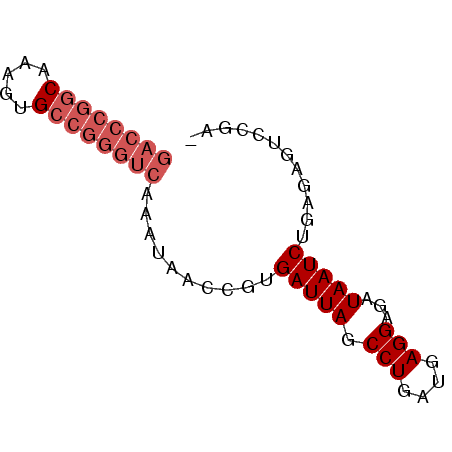
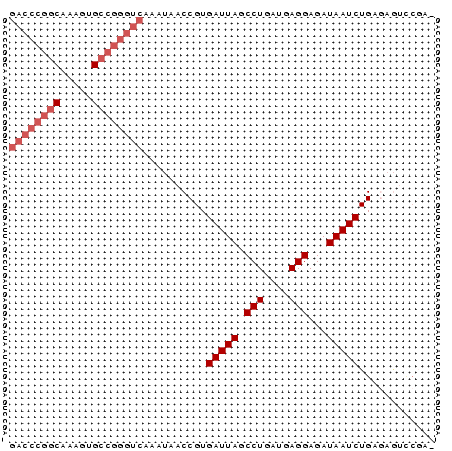
>pao_1 5284038 66 - 1 GACCCGGCAAAGUGCCGGGUCAAA-AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAAGAGUCCGA ((((((((.....))))))))...-......(((((.(((....)))...)))))............ ( -24.40) >pel_1 1 65 - 1 GACCCGGCAAGUUGCCGGGUC-AAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAGUCCGA- ((((((((.....))))))))-.........(((((.(((....)))...)))))...........- ( -24.40) >pf5_1 1 65 - 1 GACCCGGC-AAGAGCCGGGUCAAAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAGUCCGA- ((((((((-....))))))))..........(((((.(((....)))...)))))...........- ( -24.10) >pfo_1 1 52 - 1 --------------CCGGGUCAAAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAGUCCGA- --------------.(((((......)))..(((((.(((....)))...)))))........)).- ( -9.00) >ppk_1 1 65 - 1 GACCCGGCAAAUUGCCGGGUC-AAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAGUCCGA- ((((((((.....))))))))-.........(((((.(((....)))...)))))...........- ( -24.40) >pst_1 1 58 - 1 GACCCGGCAAAGAGCCGGGUCAAA-AACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAA-------- ((((((((.....))))))))...-......(((((.(((....)))...)))))....-------- ( -24.10) >consensus GACCCGGCAAAGUGCCGGGUCAAAUAACCGUGAUUAGCCUGAUGAGGAGAUAAUCUGAGAGUCCGA_ ((((((((.....))))))))..........(((((.(((....)))...)))))............ (-18.23 = -19.43 + 1.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:53:16 2007