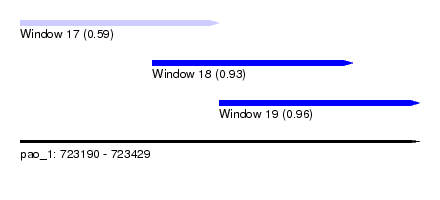
| Sequence ID | pao_1 |
|---|---|
| Location | 723,190 – 723,429 |
| Length | 239 |
| Max. P | 0.957513 |

| Location | 723,190 – 723,309 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -44.75 |
| Energy contribution | -44.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723190 119 + 1 AACGAGCGCAACCCUUGUCCUUAGUUACCAGCACCUCG-GGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCA ...........(((...(((((.((((((..((((...-))))((((.(((....))).))))))))))......)))))....)))((((((.....)))))).(((((.....))))) ( -46.40) >pf5_1 1001 120 + 1 AACGAGCGCAACCCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCU ...........(((...(((((.((((((..(((......)))((((.(((....))).))))))))))......)))))....)))((((((.....))))))..((((.....)))). ( -44.30) >psp_1 1001 120 + 1 AACGAGCGCAACCCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCU ...........(((...(((((.((((((..(((......)))((((.(((....))).))))))))))......)))))....)))((((((.....))))))..((((.....)))). ( -44.30) >pss_1 1001 120 + 1 AACGAGCGCAACCCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCU ...........(((...(((((.((((((..(((......)))((((.(((....))).))))))))))......)))))....)))((((((.....))))))..((((.....)))). ( -44.30) >consensus AACGAGCGCAACCCUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCU ...........(((...(((((.((((((..(((......)))((((.(((....))).))))))))))......)))))....)))((((((.....))))))..((((.....)))). (-44.75 = -44.75 + 0.00)

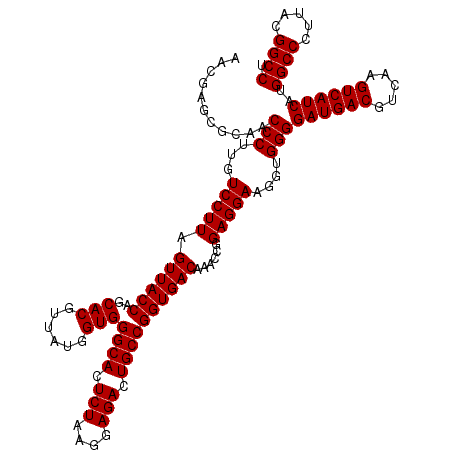
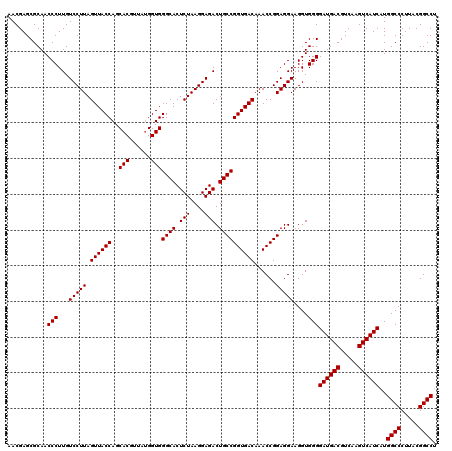
| Location | 723,269 – 723,389 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -46.22 |
| Consensus MFE | -45.42 |
| Energy contribution | -45.18 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.930848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723269 120 + 1 AGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUA ..((((.((((((.....)))))).(((((((.......)))))))........)))).(((((((((.....(((..((...(((....)))...))....))).....))))))))). ( -47.10) >pf5_1 1081 120 + 1 AGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUA ..((((.((((((.....)))))).((((((.........))))))........)))).(((((((((.....(((..((...(((....)))...))....))).....))))))))). ( -46.40) >psp_1 1081 120 + 1 AGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUA ..((((.((((((.....)))))).((((((.........))))))........)))).(((((((((...((((...((...(((....)))...))...)))).....))))))))). ( -45.70) >pss_1 1081 120 + 1 AGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUA ..((((.((((((.....)))))).((((((.........))))))........)))).(((((((((...((((...((...(((....)))...))...)))).....))))))))). ( -45.70) >consensus AGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUA ..((((.((((((.....)))))).((((((.........))))))........)))).(((((((((.....(((((((...(((....)))...)).)))))......))))))))). (-45.42 = -45.18 + -0.25)

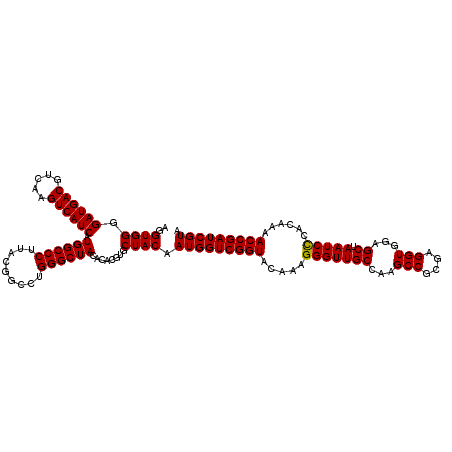
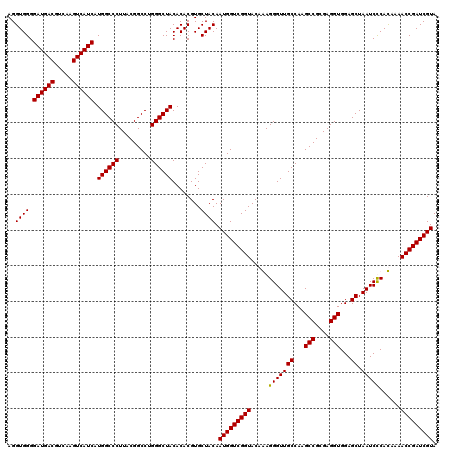
| Location | 723,309 – 723,429 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -48.25 |
| Consensus MFE | -47.70 |
| Energy contribution | -47.45 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723309 120 + 1 GGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGA .((((.(((.((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..))....((((((........))))))))).))))... ( -48.60) >pf5_1 1121 120 + 1 GGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGA .((((.(((.((..((((...(((((((.....(((..((...(((....)))...))....))).....)))))))))))..))....((((((........))))))))).))))... ( -48.60) >psp_1 1121 120 + 1 GGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGA .((((.(((.((..((((...(((((((...((((...((...(((....)))...))...)))).....)))))))))))..))....((((((........))))))))).))))... ( -47.90) >pss_1 1121 120 + 1 GGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGA .((((.(((.((..((((...(((((((...((((...((...(((....)))...))...)))).....)))))))))))..))....((((((........))))))))).))))... ( -47.90) >consensus GGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGA .((((.(((.((..((((...(((((((.....(((((((...(((....)))...)).)))))......)))))))))))..))....((((((........))))))))).))))... (-47.70 = -47.45 + -0.25)

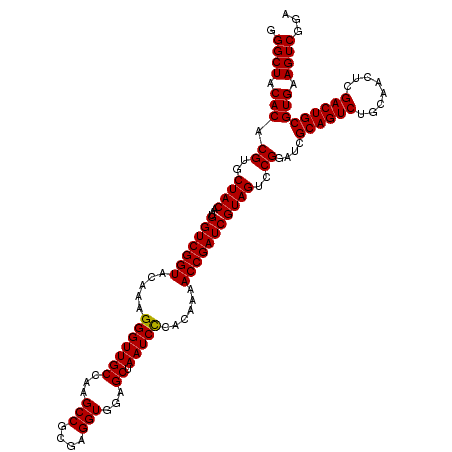
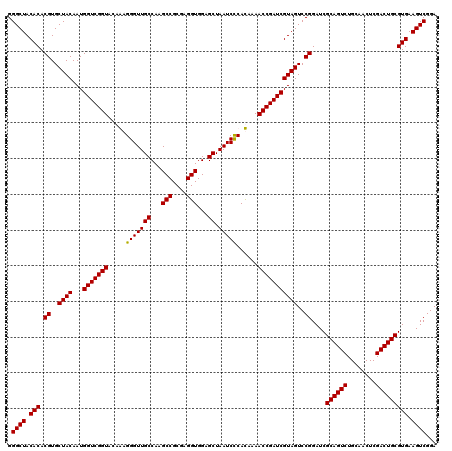
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:45:51 2007