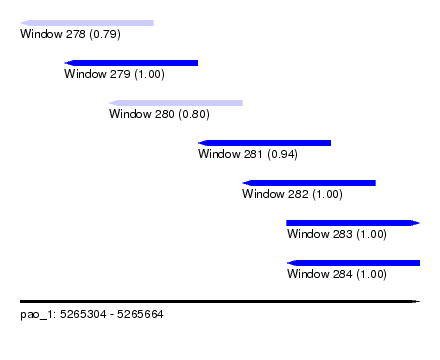
| Sequence ID | pao_1 |
|---|---|
| Location | 5,265,304 – 5,265,664 |
| Length | 360 |
| Max. P | 0.999933 |

| Location | 5,265,304 – 5,265,424 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -38.80 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.75 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265304 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -39.70) >pf5_1 958 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -37.90) >pfo_1 961 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -37.90) >pfs_1 961 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -37.90) >pss_1 958 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -39.70) >pst_1 958 120 - 1 CGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. ( -39.70) >consensus CGGAAGGUUAAUUGAUGGGGUUAGCGAAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCU .((((...........((((((.((....))..))))))...(((..((((((.....))).((((((.......))))))....)))..)))..))))..((((((.....)))))).. (-38.80 = -38.80 + 0.00)

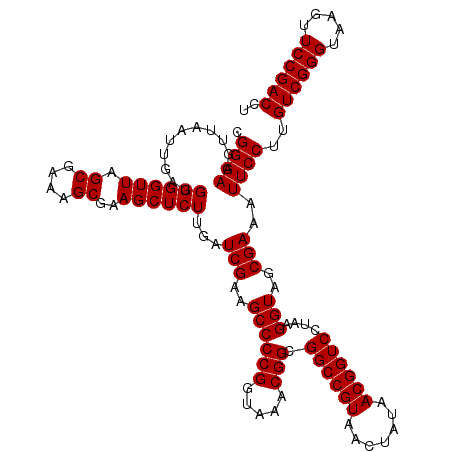
| Location | 5,265,344 – 5,265,464 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -47.60 |
| Consensus MFE | -47.60 |
| Energy contribution | -47.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265344 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -48.50) >pf5_1 998 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -46.70) >pfo_1 1001 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -46.70) >pfs_1 1001 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -46.70) >pss_1 998 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -48.50) >pst_1 998 120 - 1 CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. ( -48.50) >consensus CACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGAAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGU (((....)))..(((....(((...(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).))).)))...))).. (-47.60 = -47.60 + 0.00)


| Location | 5,265,384 – 5,265,504 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -38.10 |
| Energy contribution | -38.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265384 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUU ((((((((...(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..)))).))))..)).... ( -40.10) >pf5_1 1038 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUU ...((((((..(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..))...)))..))).... ( -37.30) >pfo_1 1041 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUU ...((((((..(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..))...)))..))).... ( -37.30) >pfs_1 1041 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUU ...((((((..(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..))...)))..))).... ( -37.30) >pss_1 1038 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUU ((((((((...(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..)))).))))..)).... ( -40.10) >pst_1 1038 120 - 1 GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUU ((((((((...(((((.....)))))..((((((((....(((....)))......))))))))...((..((((.(((((....)))......)).))))..)))).))))..)).... ( -40.10) >consensus GCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGAAAGCGAAGCUCUU (((((.(((..(((((.....)))))..((((((((....(((....)))......))))))))..)))..))..)))(((....)))........((((((.((....))..)))))). (-38.10 = -38.10 + 0.00)

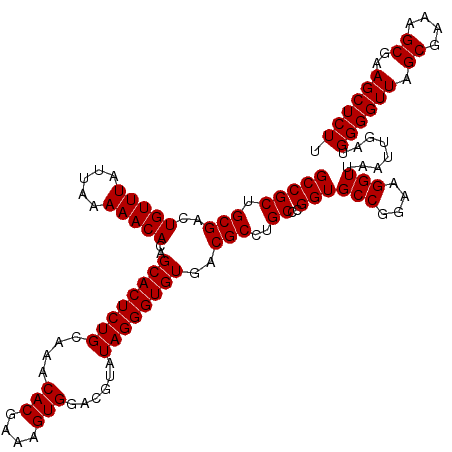
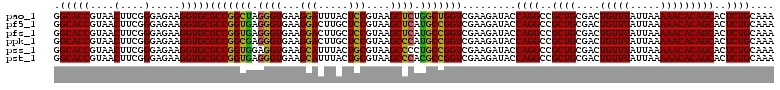
| Location | 5,265,464 – 5,265,584 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -40.83 |
| Energy contribution | -39.55 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.99 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265464 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........((((..((((....(((((.....)))))))))..)))).... ( -43.10) >pf5_1 1118 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........((((..((((....(((((.....)))))))))..)))).... ( -37.70) >pfs_1 1121 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........((((..((((....(((((.....)))))))))..)))).... ( -37.70) >ppk_1 1121 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........((((..((((....(((((.....)))))))))..)))).... ( -43.30) >pss_1 1118 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..))))).........((((..((((....(((((.....)))))))))..)))).... ( -40.30) >pst_1 1118 120 - 1 GGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........((((..((((....(((((.....)))))))))..)))).... ( -43.00) >consensus GGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAA .(((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........((((..((((....(((((.....)))))))))..)))).... (-40.83 = -39.55 + -1.28)
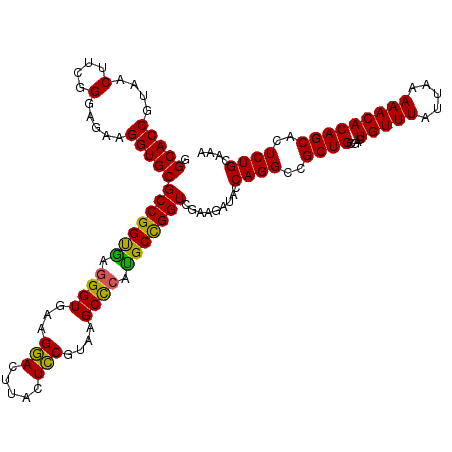
| Location | 5,265,504 – 5,265,624 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -43.45 |
| Consensus MFE | -43.42 |
| Energy contribution | -42.15 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265504 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))((((((((((((...(((.....)))....))))))))))))........)).. ( -45.70) >pf5_1 1158 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........)).. ( -40.30) >pfs_1 1161 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........)).. ( -40.30) >ppk_1 1161 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........)).. ( -45.90) >pss_1 1158 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..)))))........)).. ( -42.90) >pst_1 1158 120 - 1 AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........)).. ( -45.60) >consensus AGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAG ...((((((((....))))))))..((..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))........)).. (-43.42 = -42.15 + -1.27)

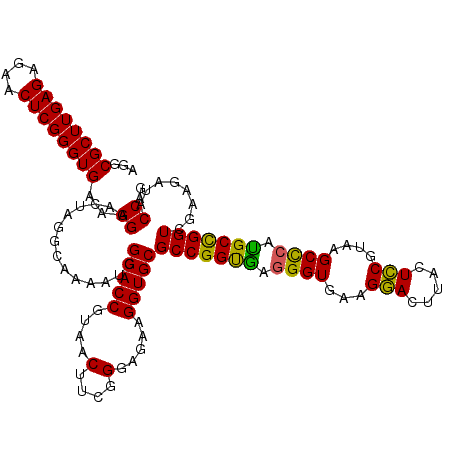
| Location | 5,265,544 – 5,265,664 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -34.55 |
| Energy contribution | -34.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.56 |
| SVM RNA-class probability | 0.999920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265544 120 + 1 UCACCCUAGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGGUAC ..((((((((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..))).........((((((....).)))))..)))))).. ( -38.60) >pel_1 1201 120 + 1 UCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. ( -34.70) >pfs_1 1201 120 + 1 UCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. ( -34.30) >ppk_1 1201 120 + 1 UCACCCUCGCCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. ( -35.00) >psp_1 1198 120 + 1 UCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. ( -34.30) >pss_1 1198 120 + 1 UCACCUCCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. ( -33.20) >consensus UCACCCUCACCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUAC ..(((((.((((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((((.....))))..............)))..))))).. (-34.55 = -34.05 + -0.50)
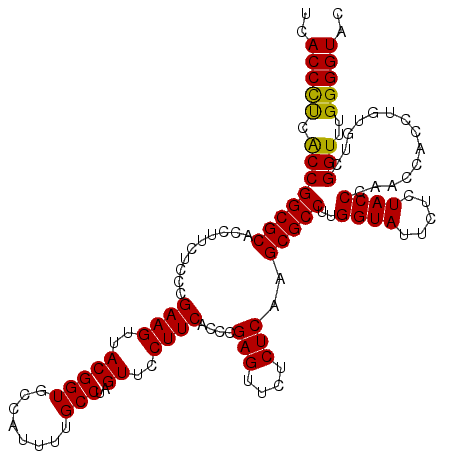
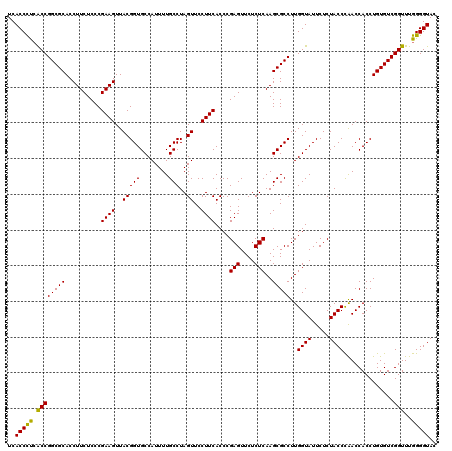
| Location | 5,265,544 – 5,265,664 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -40.53 |
| Consensus MFE | -38.68 |
| Energy contribution | -39.18 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5265544 120 - 1 GUACCCCAAACCGACACAGGUGGUCGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGA .(((((...(((......)))((((((((..((....((....((((((((....))))))))..))..))..))......(((((....(....).....))))).)))))).))))). ( -41.70) >pel_1 1201 120 - 1 GUACCCCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGA .(((((...((((.(.((..((.(((((((.....))))....((((((((....)))))))).......))).))...))(((((....(....).....)))))).))))..))))). ( -41.10) >pfs_1 1201 120 - 1 GUACCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGA .(((((...(((........(((((.((((.....))))....((((((((....))))))))....)))))(((......(((((....(....).....)))))))))))..))))). ( -40.90) >ppk_1 1201 120 - 1 GUACCCCAAACCGACACAGGUGGUUAGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGA .((((((..(((......))).(((.((((.....))))..((((((((((....)))))))...................(((((....(....).....)))))))))))).))))). ( -39.00) >psp_1 1198 120 - 1 GUACCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGA .(((((...(((........(((((.((((.....))))....((((((((....))))))))....)))))(((......(((((....(....).....)))))))))))..))))). ( -40.90) >pss_1 1198 120 - 1 GUACCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGA .(((((((..((..((((((((.((.((((.....)))).)).)))))(((....)))..)))..)).....(((......(((((....(....).....))))))))..))).)))). ( -39.60) >consensus GUACCCCAAACCGACACAGGUGGUUGGGUAGAGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGA .(((((...((((.(...((((....((((.....))))....((((((((....))))))))...................))))(((.((((....)))).)))).))))..))))). (-38.68 = -39.18 + 0.50)
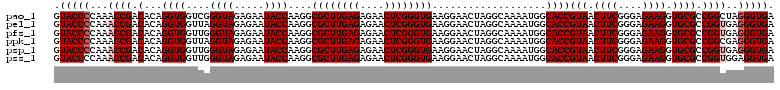
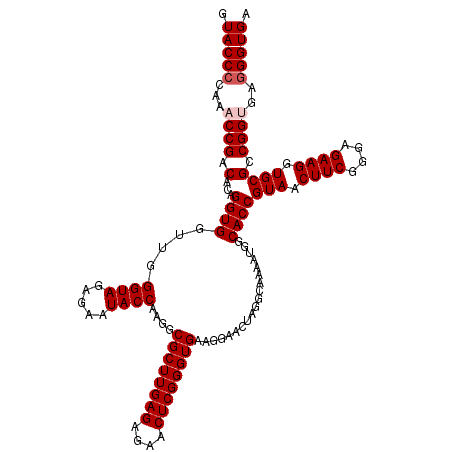
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:51:45 2007