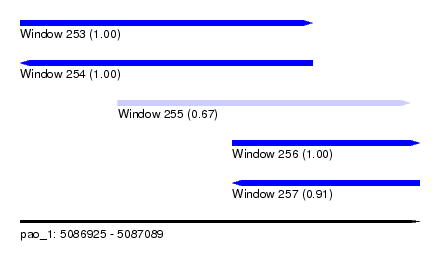
| Sequence ID | pao_1 |
|---|---|
| Location | 5,086,925 – 5,087,089 |
| Length | 164 |
| Max. P | 0.999449 |

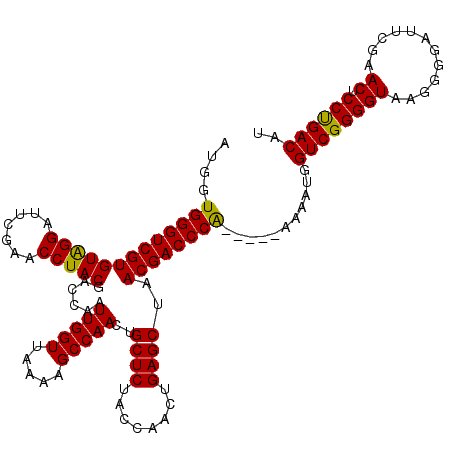
| Location | 5,086,925 – 5,087,045 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -39.00 |
| Energy contribution | -38.33 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
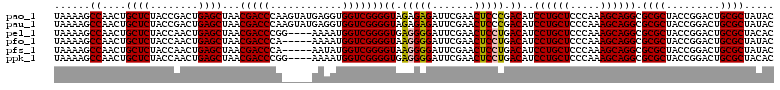
>pao_1 5086925 120 + 1 AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCCAAGUAUGAGGUGGUCGGGGUAGAGAGAUUCGAACUCCCGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..))))))))...........((((((((.(((...)))..)).)))))).. ( -41.30) >pau_1 1 120 + 1 AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCCAAGUAUGAGGUGGUCGGGGUAGAGAGAUUCGAACUCCCGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..))))))))...........((((((((.(((...)))..)).)))))).. ( -41.30) >pel_1 1 116 + 1 AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGG----AAAAUGGUCGGGGUGAGGGGAUUCGAACUCCUGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))).----......((((((((..((....))..)).)))))).. ( -39.30) >pfo_1 1 115 + 1 AUGGUGGGUCGUGUGGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA-----AAAAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..))))))))-----......((((((((..((....))..)).)))))).. ( -38.70) >pfs_1 1 115 + 1 AUGGUGGGUCGUGUGGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA-----AAUAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..))))))))-----......((((((((..((....))..)).)))))).. ( -38.70) >ppk_1 1 116 + 1 AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGG----AAAAUGGUCGGGGUGAGGGGAUUCGAACUCCUGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..)))))))).----......((((((((..((....))..)).)))))).. ( -39.30) >consensus AUGGUGGGUCGUGUAGGAUUCGAACCUACGACCAAUUGGUUAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA_____AAAAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAU ....(((((((((((((.......)))))......(((((.....)))))..((((........))))..))))))))...........((((((((............)).)))))).. (-39.00 = -38.33 + -0.66)

| Location | 5,086,925 – 5,087,045 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5086925 120 - 1 AUGUCGGGAGUUCGAAUCUCUCUACCCCGACCACCUCAUACUUGGGUCGUUAGCUCAGUCGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU ..((((((.((..((.....)).))))))))...........((((((((..............((.(((((((.......))))))).))(((((.......))))))))))))).... ( -41.70) >pau_1 1 120 - 1 AUGUCGGGAGUUCGAAUCUCUCUACCCCGACCACCUCAUACUUGGGUCGUUAGCUCAGUCGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU ..((((((.((..((.....)).))))))))...........((((((((..............((.(((((((.......))))))).))(((((.......))))))))))))).... ( -41.70) >pel_1 1 116 - 1 AUGUCAGGAGUUCGAAUCCCCUCACCCCGACCAUUUU----CCGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU ..(((.((.((..((......)))))).)))......----..(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......))))))))))))..... ( -38.20) >pfo_1 1 115 - 1 AUGUCAGGAGUUCGAAUCCCCUUACCCCGACCAUUUU-----UGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCCACACGACCCACCAU ..(((.((..((((((((..(......(((((.....-----..)))))...(..(((((((((((((.....)))))..))))))))..))..))))))))..))....)))....... ( -35.90) >pfs_1 1 115 - 1 AUGUCAGGAGUUCGAAUCCCCUUACCCCGACCAUAUU-----UGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCCACACGACCCACCAU ..(((.((..((((((((..(......(((((.....-----..)))))...(..(((((((((((((.....)))))..))))))))..))..))))))))..))....)))....... ( -35.90) >ppk_1 1 116 - 1 AUGUCAGGAGUUCGAAUCCCCUCACCCCGACCAUUUU----CCGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU ..(((.((.((..((......)))))).)))......----..(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......))))))))))))..... ( -38.20) >consensus AUGUCAGGAGUUCGAAUCCCCUUACCCCGACCAUUUU_____UGGGUCGUUAGCUCAGUUGGUAGAGCAGUUGGCUUUUAACCAAUUGGUCGUAGGUUCGAAUCCUACACGACCCACCAU ..(((.((.((..(......)..)))).)))............(((((((..(..(((((((((((((.....)))))..))))))))..)(((((.......))))))))))))..... (-35.26 = -35.70 + 0.44)

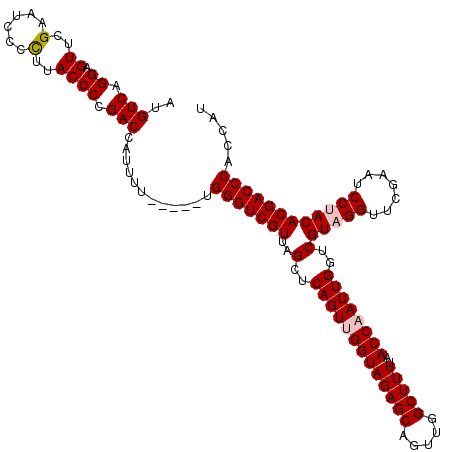
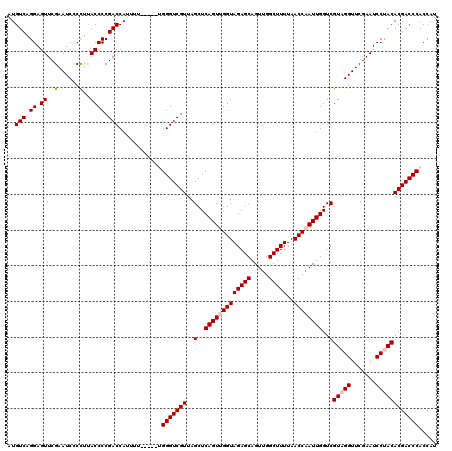
| Location | 5,086,965 – 5,087,085 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -34.13 |
| Energy contribution | -33.80 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5086965 120 + 1 UAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCCAAGUAUGAGGUGGUCGGGGUAGAGAGAUUCGAACUCCCGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUAC .....(((....((((........))))..((.(((((....)).))).)).(((((((.((...(((......)))..)))))).)))......)))((((.........))))..... ( -36.10) >pau_1 41 120 + 1 UAAAAGCCAACUGCUCUACCGACUGAGCUAACGACCCAAGUAUGAGGUGGUCGGGGUAGAGAGAUUCGAACUCCCGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUAC .....(((....((((........))))..((.(((((....)).))).)).(((((((.((...(((......)))..)))))).)))......)))((((.........))))..... ( -36.10) >pel_1 41 116 + 1 UAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGG----AAAAUGGUCGGGGUGAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUACAC ......((....((((........))))...(((((...----.....)))))))((.(((((.......))))).))..((((((.....)))))).((((.........))))..... ( -37.60) >pfo_1 41 115 + 1 UAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA-----AAAAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUAC ......((....((((........))))...(((((..-----.....)))))))((.(((((.......))))).))..((((((.....)))))).((((.........))))..... ( -35.20) >pfs_1 41 115 + 1 UAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA-----AAUAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUAC ......((....((((........))))...(((((..-----.....)))))))((.(((((.......))))).))..((((((.....)))))).((((.........))))..... ( -35.20) >ppk_1 41 116 + 1 UAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCGG----AAAAUGGUCGGGGUGAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUACAC ......((....((((........))))...(((((...----.....)))))))((.(((((.......))))).))..((((((.....)))))).((((.........))))..... ( -37.60) >consensus UAAAAGCCAACUGCUCUACCAACUGAGCUAACGACCCA_____AAAAUGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUAC ......((....((((........))))...(((((............)))))))((.(((((.......))))).))..((((((.....)))))).((((.........))))..... (-34.13 = -33.80 + -0.33)
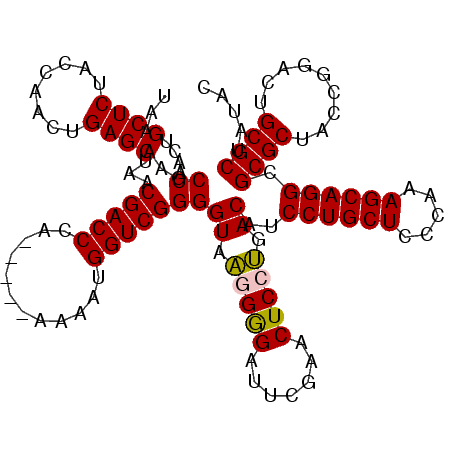
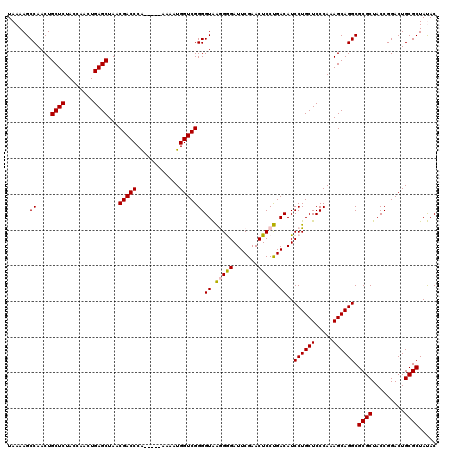
| Location | 5,087,012 – 5,087,089 |
|---|---|
| Length | 77 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.52 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5087012 77 + 1 UGGUCGGGGUAGAGAGAUUCGAACUCCCGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((.(((.......))).).....((((((.....)))))).((((.........))))..))))))) ( -27.20) >pf5_1 1 77 + 1 UGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) ( -30.60) >pfo_1 1 77 + 1 UGGUCGGGGUAGGGGGAUUCGAACUCCCGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) ( -33.20) >psp_1 1 77 + 1 UGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) ( -30.60) >pss_1 1 77 + 1 UGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) ( -30.60) >pst_1 1 77 + 1 UGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) ( -30.60) >consensus UGGUCGGGGUAAGGGGAUUCGAACUCCUGACAUCCUGCUCCCAAAGCAGGCGCGCUACCGGACUGCGCUAUACCCCG ....((((((((((((.......))))).....((((((.....)))))).((((.........))))..))))))) (-30.93 = -30.52 + -0.42)


| Location | 5,087,012 – 5,087,089 |
|---|---|
| Length | 77 |
| Sequences | 6 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 97.75 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.27 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5087012 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCGGGAGUUCGAAUCUCUCUACCCCGACCA (((((((..((((.........)))).(((((((...))))))).....(((((.......)))))))))))).... ( -32.90) >pf5_1 1 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCAGGAGUUCGAAUCCCCUUACCCCGACCA (((((((..((((.........)))).(((((((...)))))))......(((.......)))...))))))).... ( -29.70) >pfo_1 1 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCGGGAGUUCGAAUCCCCCUACCCCGACCA (((((((..((((.........)))).(((((((...))))))).....((((.......))))..))))))).... ( -33.00) >psp_1 1 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCAGGAGUUCGAAUCCCCUUACCCCGACCA (((((((..((((.........)))).(((((((...)))))))......(((.......)))...))))))).... ( -29.70) >pss_1 1 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCAGGAGUUCGAAUCCCCUUACCCCGACCA (((((((..((((.........)))).(((((((...)))))))......(((.......)))...))))))).... ( -29.70) >pst_1 1 77 - 1 CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCAGGAGUUCGAAUCCCCUUACCCCGACCA (((((((..((((.........)))).(((((((...)))))))......(((.......)))...))))))).... ( -29.70) >consensus CGGGGUAUAGCGCAGUCCGGUAGCGCGCCUGCUUUGGGAGCAGGAUGUCAGGAGUUCGAAUCCCCUUACCCCGACCA (((((((..((((.........)))).(((((((...)))))))......(((.......)))...))))))).... (-29.41 = -29.27 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:51:08 2007