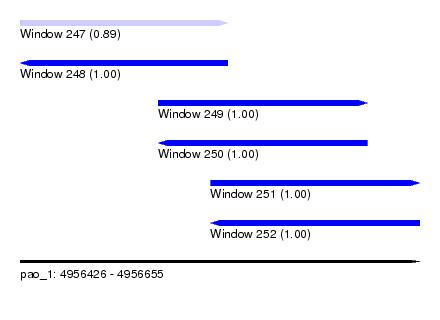
| Sequence ID | pao_1 |
|---|---|
| Location | 4,956,426 – 4,956,655 |
| Length | 229 |
| Max. P | 0.999997 |

| Location | 4,956,426 – 4,956,545 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -44.13 |
| Consensus MFE | -39.29 |
| Energy contribution | -37.85 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
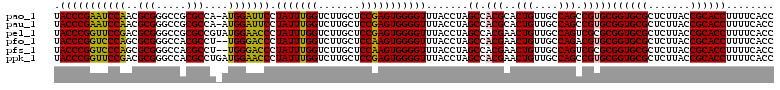
>pao_1 4956426 119 + 1 UACCCGAAUCCAACGCGGGCCGCGCCA-AUGGAUUCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGCACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACC .....(((((((..(((.....)))..-.)))))))((((((((.......))))))))((((.....))))..((((.((.....)).))))((((((.......))))))........ ( -40.00) >pau_1 81 119 + 1 UACCCGAAUCCAACGCGGGCCGCGCCA-AUGGAUUCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGCACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACC .....(((((((..(((.....)))..-.)))))))((((((((.......))))))))((((.....))))..((((.((.....)).))))((((((.......))))))........ ( -40.00) >pel_1 81 120 + 1 UACCCGGUUCCGACGCGGGCCGCGCCGUAUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGUCGCGCGGUGCGCUCUUACCGCACCUUUUCACC ....((((...((.(((.(((((((......(((((((((((((.......)))))))))))))..........((.(((....)))))))))))).)))))..))))............ ( -48.40) >pfo_1 81 118 + 1 UACCCGGUCCCAGCGCGGGCCACGCCU--UGGGACCCUAUUUGGUCUUGCUCCAAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCACC .....((((((((.(((.....))).)--)))))))((((((((.......))))))))((((.....))))..((((((....)))..((((((((......))))))))...)))... ( -46.30) >pfs_1 81 118 + 1 UACCCGGUCCCAGCGCGGGCCACGCCU--UGGGACCCUAUUUGGUCUUGCUCCAAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGUCGCGCGGUGCGCUCUUACCGCACCUUUUCACC .((((((((((((.(((.....))).)--))))))).(((((((.......))))))))))).......((..(((.(((....)))))).))((((((.......))))))........ ( -46.40) >ppk_1 81 120 + 1 UACCCGGUUCCGACGCGGGCCACGCCUGAUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACC .....((((.((((.(((((...)))))...(((((((((((((.......)))))))))))))...............))))..))))((((((((......))))))))......... ( -43.70) >consensus UACCCGGUUCCAACGCGGGCCACGCCU_AUGGAACCCUAUUUGGUCUUGCUCCGAGUGGGGUUUACCUAGCCACGAACUGUUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACC .(((((((((((..(((.....)))....))))))).(((((((.......))))))))))).......((.(((..(((....))).)))))((((((.......))))))........ (-39.29 = -37.85 + -1.44)
| Location | 4,956,426 – 4,956,545 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.41 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -47.36 |
| Energy contribution | -45.12 |
| Covariance contribution | -2.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.40 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4956426 119 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUGCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGAAUCCAU-UGGCGCGGCCCGCGUUGGAUUCGGGUA (((.....((((((.......)))))).((((((((....)))).))))(((.((....))((....)))))...)))......(((((((.-..(((((...))))))))))))..... ( -44.60) >pau_1 81 119 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUGCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGAAUCCAU-UGGCGCGGCCCGCGUUGGAUUCGGGUA (((.....((((((.......)))))).((((((((....)))).))))(((.((....))((....)))))...)))......(((((((.-..(((((...))))))))))))..... ( -44.60) >pel_1 81 120 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCGCGACUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAUACGGCGCGGCCCGCGUCGGAACCGGGUA (((.....((((((.......)))))).((((((((....))).))))).........)))..((((((..(..((((......)))).....(((((((...))))))))..)))))). ( -47.50) >pfo_1 81 118 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCACGUCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUUGGAGCAAGACCAAAUAGGGUCCCA--AGGCGUGGCCCGCGCUGGGACCGGGUA (((.....((((((.......)))))).((((.(((....)))..)))))))......((((...((((.......))))....(((((((--..(((((...)))))))))))))))). ( -48.80) >pfs_1 81 118 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCGCGACUGGCAACAGUUCGUGGCUAGGUAAACCCCACUUGGAGCAAGACCAAAUAGGGUCCCA--AGGCGUGGCCCGCGCUGGGACCGGGUA (((.....((((((.......)))))).((((((((....))).))))))))......((((...((((.......))))....(((((((--..(((((...)))))))))))))))). ( -50.40) >ppk_1 81 120 - 1 GGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAUCAGGCGUGGCCCGCGUCGGAACCGGGUA (((.....((((((.......)))))).((((((((....))).)))))(((.((....))((....)))))...)))......((((((....((((((...))))))))))))..... ( -44.70) >consensus GGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAACAGUUCGUGGCUAGGUAAACCCCACUCGGAGCAAGACCAAAUAGGGUUCCAU_AGGCGCGGCCCGCGUUGGAACCGGGUA (((.....((((((.......)))))).((((((((....)))).))))(((.((....))((....)))))...)))......((((((....((((((...))))))))))))..... (-47.36 = -45.12 + -2.25)

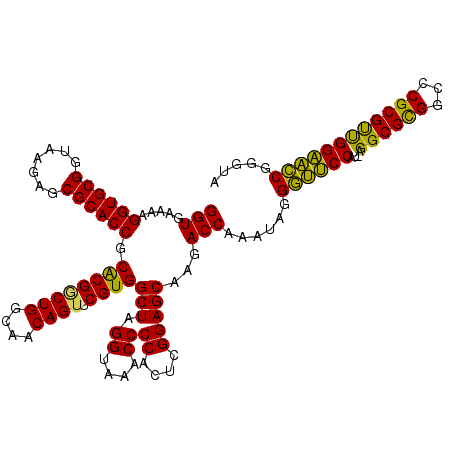
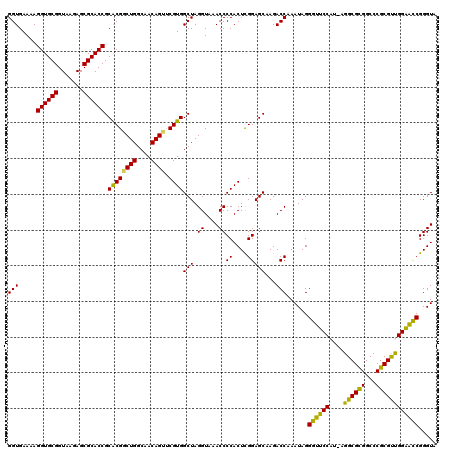
| Location | 4,956,505 – 4,956,625 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -40.46 |
| Energy contribution | -40.10 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4956505 120 + 1 UUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCA .((((.(((((((((((......))))))))......(((((((..(((((....))))))))).)))..........))).........(((...(((((.....)))))..))))))) ( -47.10) >pst_1 159 120 + 1 UUGCCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGUGUUACCUGGCA .((((((..((((((((......))))))))...............((((((((((((((...((((.......))))))))))))..(.((((....)))).)..))))))..)))))) ( -46.40) >pau_1 160 120 + 1 UUGCCAGCCGUGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCA .((((.(((((((((((......))))))))......(((((((..(((((....))))))))).)))..........))).........(((...(((((.....)))))..))))))) ( -47.10) >pel_1 161 120 + 1 UUGCCAGUCGCGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCGCCGAAGCGCUUAGGCGGUUGUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCA .((((.(((((..((((((.......)))))).....(((((((..(((((....))))))))).)))........))))).........(((...(((((.....)))))..))))))) ( -44.50) >pf5_1 159 120 + 1 UUACCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCCCCCAGGCAUUACCUGGCA ...((((..((((((((......))))))))........(((...(((....((((((((...((((.......))))))))))))))).))).....(((.....))).....)))).. ( -37.90) >pfs_1 159 120 + 1 UUGCCAGUCGCGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCUCCCAGGCAUUACCUGGCA .((((((..(.((((((((.......))))))........((...(((....((((((((...((((.......))))))))))))))).)))).)..(((.....))).....)))))) ( -40.10) >consensus UUGCCAGACGUGCGGUGCGCUCUUACCGCACCUUUUCACCCUUACCGGCACCGAAGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCA .((((((..(.((((((((.......)))))).....(((((((..(((((....))))))))).)))......((((((......)))))))).)(((((.....)))))...)))))) (-40.46 = -40.10 + -0.36)
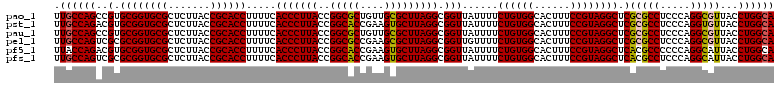
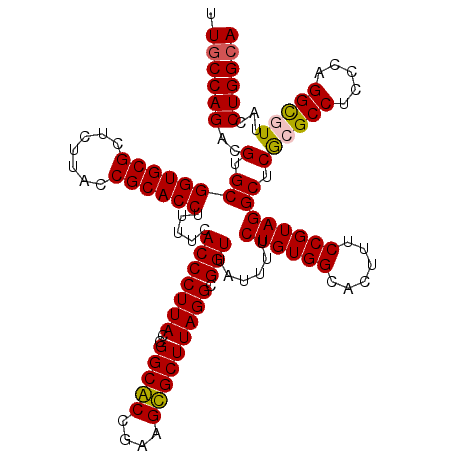
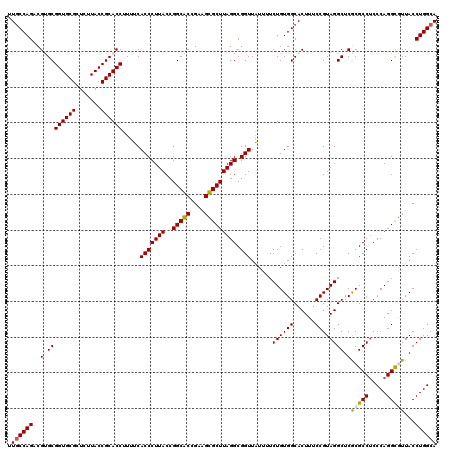
| Location | 4,956,505 – 4,956,625 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -51.08 |
| Consensus MFE | -51.72 |
| Energy contribution | -50.72 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.26 |
| Structure conservation index | 1.01 |
| SVM decision value | 6.19 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4956505 120 - 1 UGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAA ((((((.(..(((((((.((((....))))..((......))...........))(((...((((....)))).)))..))))).....(((((((........)))))))).)))))). ( -49.60) >pst_1 159 120 - 1 UGCCAGGUAACACCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCACGUCUGGCAA (((((((...(((((((.((((....))))..((......))...........))(((...((((....)))).)))..))))).....(((((((........))))))).))))))). ( -52.10) >pau_1 160 120 - 1 UGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCACGGCUGGCAA ((((((.(..(((((((.((((....))))..((......))...........))(((...((((....)))).)))..))))).....(((((((........)))))))).)))))). ( -49.60) >pel_1 161 120 - 1 UGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAACAACCGCCUAAGCGCUUCGGCGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCGCGACUGGCAA ((((((....(((((((.((((....))))..((......))...........))(((...((((....)))).)))..))))).....(((((((........)))))))..)))))). ( -52.80) >pf5_1 159 120 - 1 UGCCAGGUAAUGCCUGGGGGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCACGUCUGGUAA ..(((((.....))))).((((....)))).((((..((((.............(((((..((((....))))......)))))....((((((.......)))))))))).)))).... ( -50.70) >pfs_1 159 120 - 1 UGCCAGGUAAUGCCUGGGAGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCGCGACUGGCAA ((((((.......(((..((((....)))).)))...((((.............(((((..((((....))))......)))))....((((((.......))))))))))..)))))). ( -51.70) >consensus UGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGCGCCGGUAAGGGUGAAAAGGUGCGGUAAGAGCGCACCGCACGACUGGCAA ((((((.......(((..((((....)))).)))...((((.............(((((..((((....))))......)))))....((((((.......))))))))))..)))))). (-51.72 = -50.72 + -1.00)

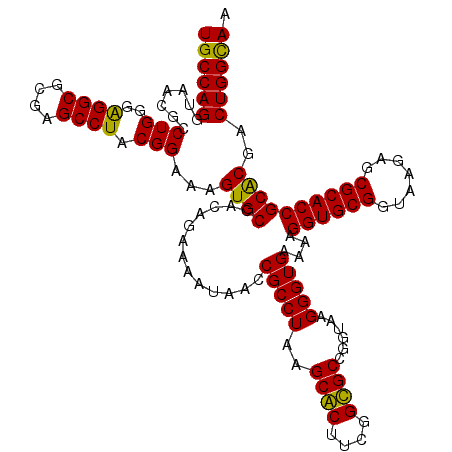
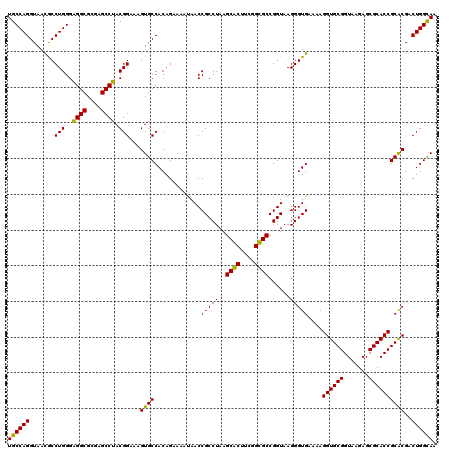
| Location | 4,956,535 – 4,956,655 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -39.38 |
| Energy contribution | -39.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4956535 120 + 1 CCUUUUCACCCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....(((((((....(((((.....))))).....(((.....)))))))))))))))))......... ( -41.20) >pst_1 189 120 + 1 CCUUUUCACCCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGUGUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....((((((((...(((....(((((.....))))).....))))))))))))))))))......... ( -40.50) >pau_1 190 120 + 1 CCUUUUCACCCUUACCGGCGCUGUUGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....(((((((....(((((.....))))).....(((.....)))))))))))))))))......... ( -41.20) >pel_1 191 120 + 1 CCUUUUCACCCUUACCGGCGCCGAAGCGCUUAGGCGGUUGUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCUGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....(((((((....(((((.....))))).....(((.....)))))))))))))))))......... ( -40.20) >pf5_1 189 120 + 1 CCUUUUCACCCUUACCGGCACCGAAGUGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCCCCCAGGCAUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....((((((((....((....(((((.....))))).....)).)))))))))))))))......... ( -36.20) >pfo_1 189 120 + 1 CCUUUUCACCCUUACCGGCGCCGAAGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCACGCCUCCCAGGCAUUACCUGGCACUUCGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....((((((((....((....(((((.....))))).....)).)))))))))))))))......... ( -37.70) >consensus CCUUUUCACCCUUACCGGCGCCGAAGCGCUUAGGCGGUUAUUUUCUGUGGCACUUUCCGUAGGCUCGCGCCUCCCAGGCGUUACCUGGCACUCCGCCCUAUGGAGCCCGGACUUUCCUCC .......(((((((..(((((....))))))))).))).....((((.(((....((((((((...(((....(((((.....))))).....))))))))))))))))))......... (-39.38 = -39.02 + -0.36)

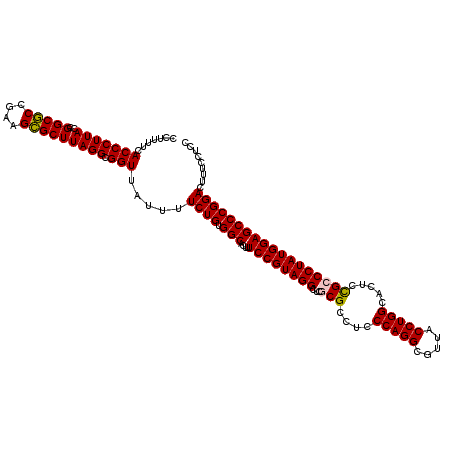
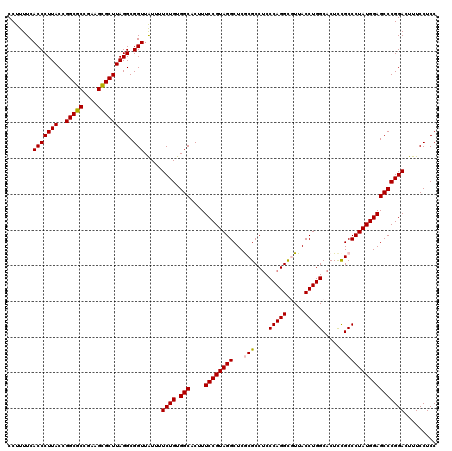
| Location | 4,956,535 – 4,956,655 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -43.40 |
| Energy contribution | -42.82 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4956535 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGGGUGAAAAGG ...((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -41.00) >pst_1 189 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCGGAGUGCCAGGUAACACCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGGGUGAAAAGG ...((....((((.(((((.......))))).(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -44.90) >pau_1 190 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCAACAGCGCCGGUAAGGGUGAAAAGG ...((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -41.00) >pel_1 191 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAACAACCGCCUAAGCGCUUCGGCGCCGGUAAGGGUGAAAAGG ...((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -44.30) >pf5_1 189 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCGGAGUGCCAGGUAAUGCCUGGGGGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCACUUCGGUGCCGGUAAGGGUGAAAAGG ...((....((((.(((((.......))))).(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -45.70) >pfo_1 189 120 - 1 GGAGGAAAGUCCGGGCUCCAUAGGGCGAAGUGCCAGGUAAUGCCUGGGAGGCGUGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCUUCGGCGCCGGUAAGGGUGAAAAGG ...((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... ( -44.80) >consensus GGAGGAAAGUCCGGGCUCCAUAGGGCAGAGUGCCAGGUAACGCCUGGGAGGCGCGAGCCUACGGAAAGUGCCACAGAAAAUAACCGCCUAAGCGCUUCGGCGCCGGUAAGGGUGAAAAGG ...((....((((.((((....))))......(((((.....))))).((((....)))).)))).....))............(((((..((((....))))......)))))...... (-43.40 = -42.82 + -0.58)

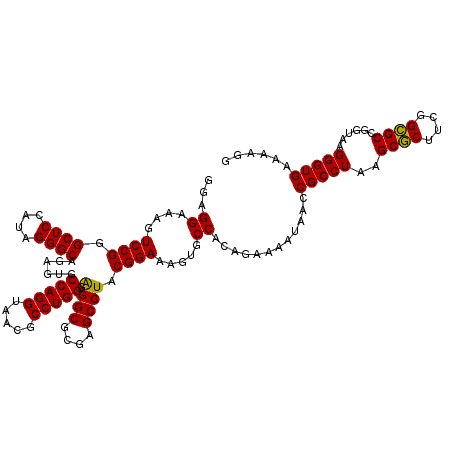
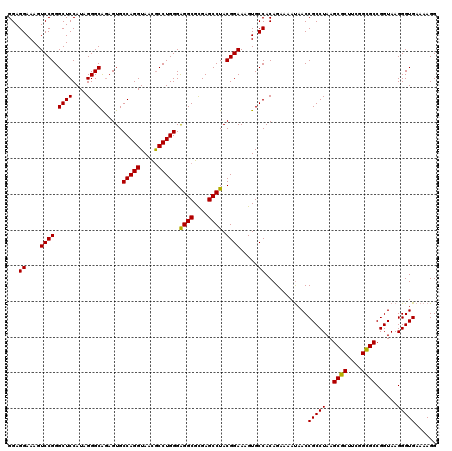
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:51:02 2007