
| Sequence ID | pao_1 |
|---|---|
| Location | 4,792,826 – 4,793,639 |
| Length | 813 |
| Max. P | 0.999994 |

| Location | 4,792,826 – 4,792,946 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -46.83 |
| Consensus MFE | -48.05 |
| Energy contribution | -46.83 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.03 |
| SVM decision value | 5.86 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
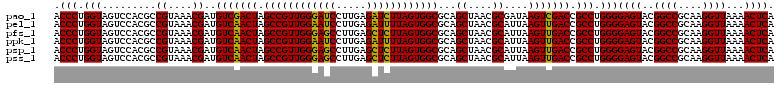
>pao_1 4792826 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAAGUCGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -48.10) >pel_1 681 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -44.00) >pfs_1 681 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -48.30) >ppk_1 681 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -44.00) >psp_1 680 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -48.30) >pss_1 680 120 - 1 ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). ( -48.30) >consensus ACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUAAGUUGACCGCCUGGGGAGUACGGCCGCAAGGUUAAAACUCA .(((.(((.........((....))..(((((((.((((((((((((.....))))))))))))...((....))....))))))).))).)))((((..((((....))))...)))). (-48.05 = -46.83 + -1.22)
| Location | 4,792,866 – 4,792,986 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -37.05 |
| Energy contribution | -36.27 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.998270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
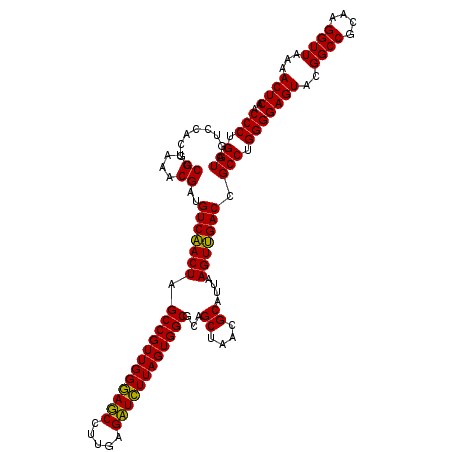
>pao_1 4792866 120 + 1 UUAUCGCGUUAGCUGCGCCACUAAGAUCUCAAGGAUCCCAACGGCUAGUCGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG .....((((..((((.(((.....((((.....)))).....)))))))..)).........(((((((.....((((((.....))))))........)))))))...))......... ( -34.12) >pel_1 721 120 + 1 UUAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG ....(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))........ ( -34.72) >pfs_1 721 120 + 1 UUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG ....(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))........ ( -38.92) >ppk_1 721 120 + 1 UUAAUGCGUUAGCUGCGCCACUAAAAUCUCAAGGAUUCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG ....(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))........ ( -34.72) >psp_1 720 120 + 1 UUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG ....(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))........ ( -38.92) >pss_1 720 120 + 1 UUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGCUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG ....(((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...)))........ ( -38.92) >consensus UUAAUGCGUUAGCUGCGCCACUAAGAGCUCAAGGAUCCCAACGGCUAGUUGACAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUG .....((((((((((.(((.....((((.....)))).....))))))))))).........(((((((.....((((((.....))))))........)))))))...))......... (-37.05 = -36.27 + -0.78)

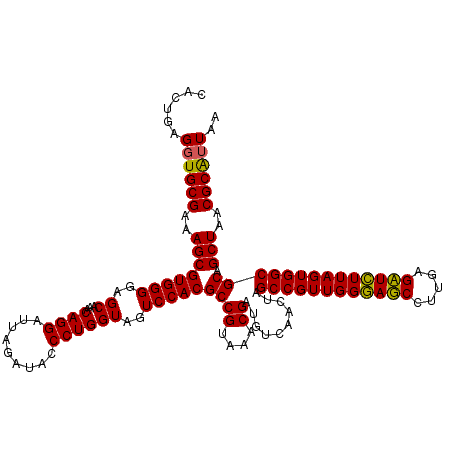
| Location | 4,792,866 – 4,792,986 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -38.55 |
| Consensus MFE | -38.81 |
| Energy contribution | -37.90 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.96 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
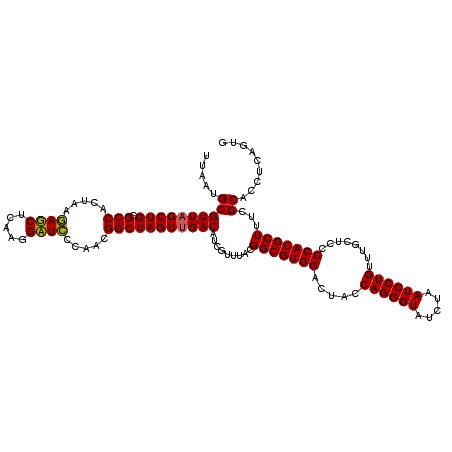
>pao_1 4792866 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCGACUAGCCGUUGGGAUCCUUGAGAUCUUAGUGGCGCAGCUAACGCGAUAA .........((....))(((...(((...((((.........)))).(((((.(((.((....)).))).)))))((((((((((((.....))))))))))))...)))..)))..... ( -39.40) >pel_1 721 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. ( -35.80) >pfs_1 721 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. ( -40.10) >ppk_1 721 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGAAUCCUUGAGAUUUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. ( -35.80) >psp_1 720 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. ( -40.10) >pss_1 720 120 - 1 CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGCUCUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. ( -40.10) >consensus CACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGUCAACUAGCCGUUGGGAGCCUUGAGAUCUUAGUGGCGCAGCUAACGCAUUAA ......((((((..((((((((..((...((((.........))))))..)))))((((....))..........((((((((((((.....)))))))))))))).)))..)))))).. (-38.81 = -37.90 + -0.91)

| Location | 4,792,946 – 4,793,066 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -25.40 |
| Energy contribution | -25.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792946 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >pfo_1 801 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >pfs_1 801 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >ppk_1 801 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >psp_1 800 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >pss_1 800 120 + 1 AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... ( -25.40) >consensus AUCUAAUCCUGUUUGCUCCCCACGCUUUCGCACCUCAGUGUCAGUAUCAGUCCAGGUGGUCGCCUUCGCCACUGGUGUUCCUUCCUAUAUCUACGCAUUUCACCGCUACACAGGAAAUUC ......((((((..((.......((....))......((((..((((......(((.((.((((.........))))..))..)))))))..))))........))...))))))..... (-25.40 = -25.40 + 0.00)

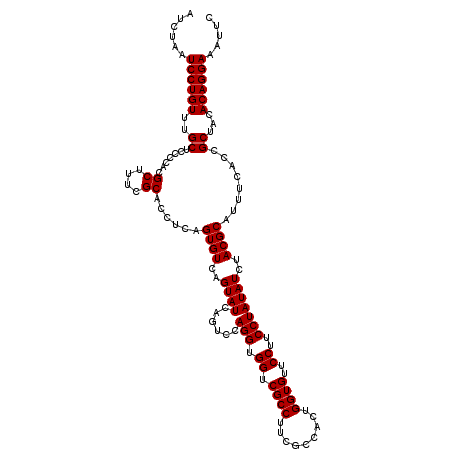
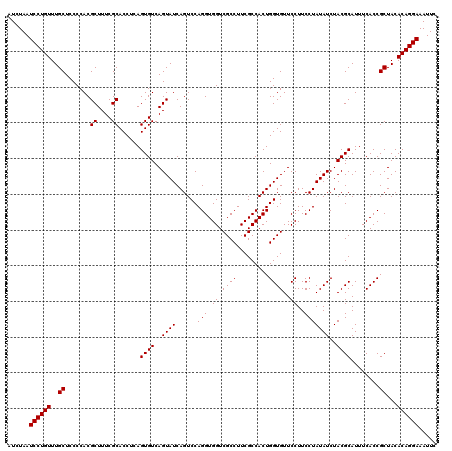
| Location | 4,792,946 – 4,793,066 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -35.50 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792946 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >pfo_1 801 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >pfs_1 801 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >ppk_1 801 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >psp_1 800 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >pss_1 800 120 - 1 GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. ( -35.50) >consensus GAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGACACUGAGGUGCGAAAGCGUGGGGAGCAAACAGGAUUAGAU ...((((((((((.(((......)))....)))))))))).....(((((((((....)).))).)))).......((((..(((....((....))((.....))...)))..)))).. (-35.50 = -35.50 + 0.00)

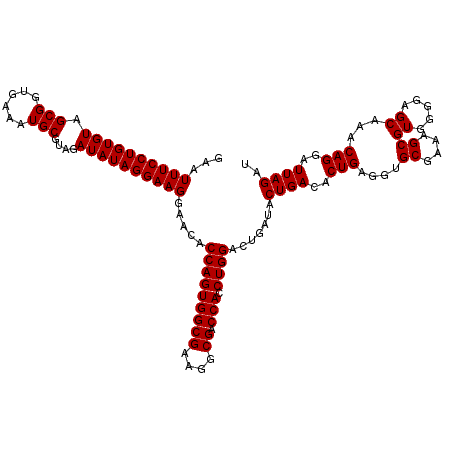
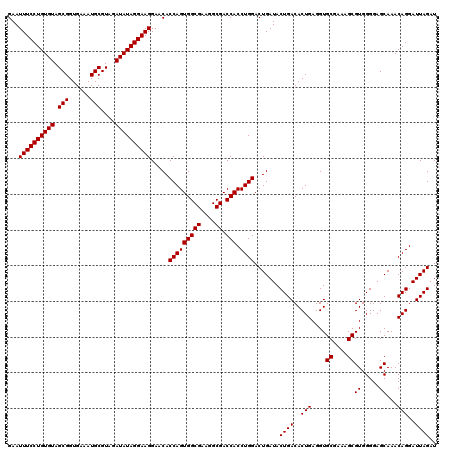
| Location | 4,792,986 – 4,793,106 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -42.23 |
| Consensus MFE | -40.71 |
| Energy contribution | -40.68 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.31 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792986 120 - 1 CAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA (((((....((((((.((....)).))))))(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))))).))....... ( -43.10) >pel_1 841 120 - 1 CAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA .........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))).. ( -42.30) >pfo_1 841 120 - 1 CAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA .........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))).. ( -42.30) >pfs_1 841 120 - 1 CAUUCAAAACUGACUGACUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA ((.(((....))).)).....((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))).. ( -39.60) >ppk_1 841 120 - 1 CAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA .........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).)))).. ( -43.80) >pss_1 840 120 - 1 CAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA .........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..))).))))).. ( -42.30) >consensus CAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAGGAACACCAGUGGCGAAGGCGACCACCUGGACUGAUACUGA .........((((....))))((((((((..(((.(((((...((((((((((.(((......)))....))))))))))...)))))((.((....)).))..)))..)))).)))).. (-40.71 = -40.68 + -0.03)

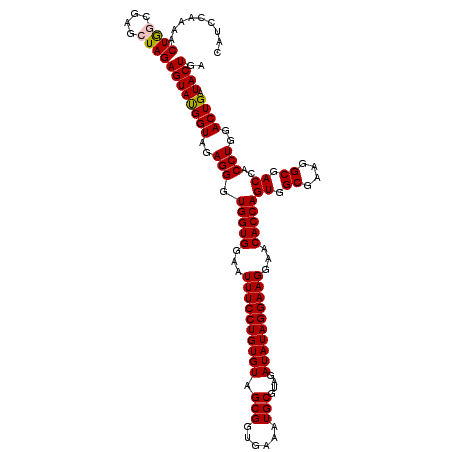
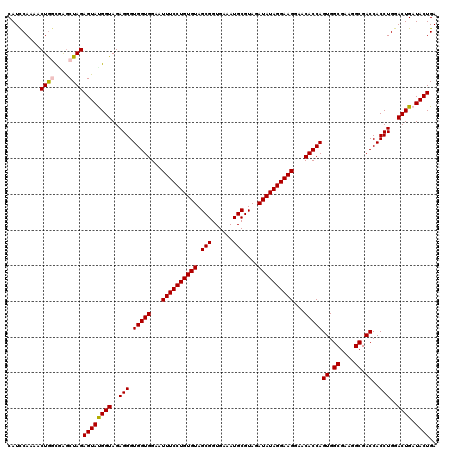
| Location | 4,793,026 – 4,793,146 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -33.95 |
| Energy contribution | -33.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793026 120 - 1 CAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ....(((((((((...((((.((......)).))))..)))))))))..((((((.((....)).)))))).............(((((((((.(((......)))....))))))))). ( -40.50) >pel_1 881 120 - 1 UAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ..((((....((....))((((((.....))))))))))...((((..(((..(..((((.....)))).)..)))..))))..(((((((((.(((......)))....))))))))). ( -36.40) >pf5_1 881 120 - 1 UAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ....((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).))).............(((((((((.(((......)))....))))))))). ( -36.80) >pfs_1 881 120 - 1 UAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUCAAAACUGACUGACUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ....(((((((((...((((.((......)).))))..))))))))).(((..((.((((.....))))...))..))).....(((((((((.(((......)))....))))))))). ( -34.80) >ppk_1 881 120 - 1 UAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ....((((((((......((((((.....))))))....))))))))..((((....))))(.(((........))).).....(((((((((.(((......)))....))))))))). ( -35.90) >pss_1 880 120 - 1 UAAGUUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ................((((((..(((.(((..(((......)))....((((....)))).....))).)))..))).)))..(((((((((.(((......)))....))))))))). ( -33.30) >consensus UAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUGGAAUUUCCUGUGUAGCGGUGAAAUGCGUAGAUAUAGGAAG ....((((((((......((((((.....))))))....))))))))..(((.((.((....)).)).))).............(((((((((.(((......)))....))))))))). (-33.95 = -33.73 + -0.22)

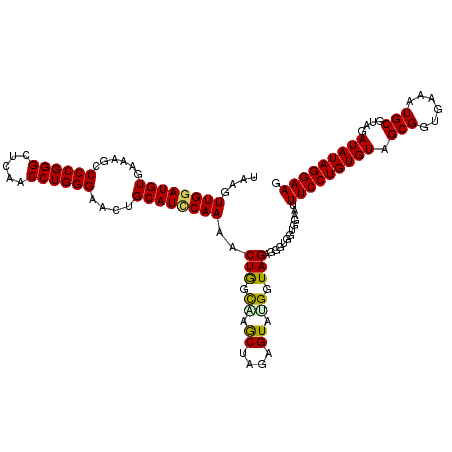
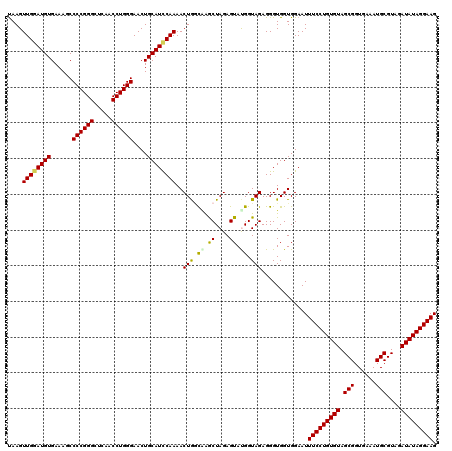
| Location | 4,793,066 – 4,793,186 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -40.19 |
| Energy contribution | -39.08 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.14 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793066 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCAGCAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUACUGAGCUAGAGUACGGUAGAGGGUGGUG ((((......(((.((((.....)))).))).((((((((....(((((((((...((((.((......)).))))..))))))))).....)))))))).....))))........... ( -41.70) >pel_1 921 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUG ......(((((((.(((.....))).).....((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).............)))))). ( -41.50) >pfo_1 921 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUG ......(((((((.(((.....))).).....((((((((((..(((((((((...((((.((......)).))))..)))))))))...)))))))))).............)))))). ( -41.70) >pfs_1 921 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUUCAAAACUGACUGACUAGAGUAUGGUAGAGGGUGGUG ......(((((((.(((.....))).).....(((((.((((..(((((((((...((((.((......)).))))..)))))))))...)))).))))).............)))))). ( -33.80) >ppk_1 921 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGGAUGUGAAAGCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUACGGUAGAGGGUGGUG ((((......(((.((((.....)))).))).((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).....))))........... ( -41.80) >pss_1 920 120 - 1 AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUUGUUAAGUUGAAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCAAGCUAGAGUAUGGUAGAGGGUGGUG ......(((((((.(((.....))).).....((((((((((..(((.(((((...((((.((......)).))))..))))).)))...)))))))))).............)))))). ( -33.10) >consensus AUCGGAAUUACUGGGCGUAAAGCGCGCGUAGGUGGUUCGUUAAGUUGGAUGUGAAAUCCCCGGGCUCAACCUGGGAACUGCAUCCAAAACUGGCGAGCUAGAGUAUGGUAGAGGGUGGUG ((((......(((.((((.....)))).))).((((((((((..((((((((......((((((.....))))))....))))))))...)))))))))).....))))........... (-40.19 = -39.08 + -1.11)

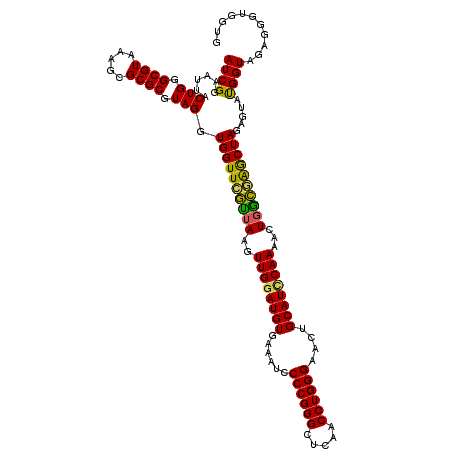
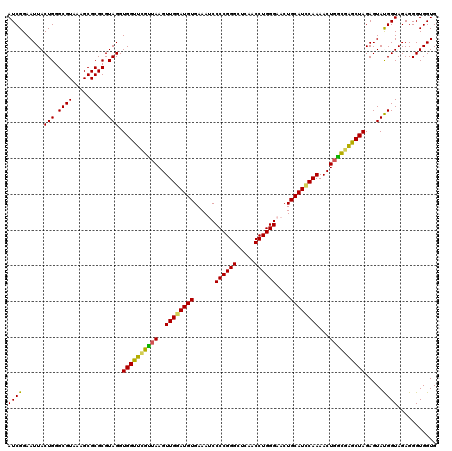
| Location | 4,793,186 – 4,793,306 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -40.94 |
| Energy contribution | -37.92 |
| Covariance contribution | -3.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.08 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793186 120 - 1 CACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUUCGUGCCAGCAGCCGCGGUAAUACGAAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -38.40) >pel_1 1041 120 - 1 CACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -39.00) >pfo_1 1041 120 - 1 CACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -37.50) >ppk_1 1041 120 - 1 CACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -39.00) >psp_1 1040 120 - 1 CACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -36.80) >pss_1 1040 120 - 1 CACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ ( -36.80) >consensus CACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUCUGUGCCAGCAGCCGCGGUAAUACAGAGGGUGCAAGCGUUA ........(((((..(..(((((((((((.......)))))))))))..)....))))).......(((((......(((((((((.((....)))))...)))))))))))........ (-40.94 = -37.92 + -3.02)
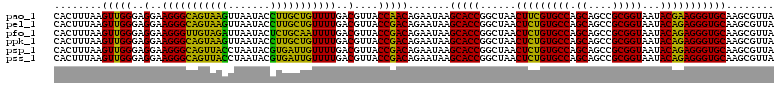
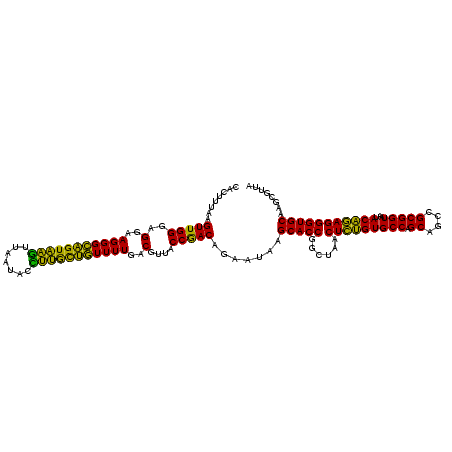
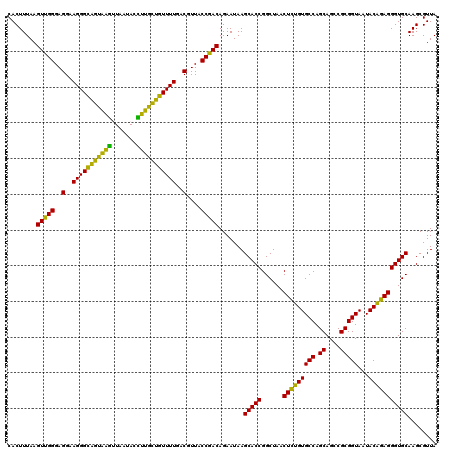
| Location | 4,793,226 – 4,793,346 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -41.53 |
| Energy contribution | -39.07 |
| Covariance contribution | -2.46 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.99 |
| Structure conservation index | 1.06 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793226 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCAACAGAAUAAGCACCGGCUAACUU .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -40.30) >pel_1 1081 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -40.00) >pfo_1 1081 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGUUGUAGAUUAAUACUCUGCAAUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -38.50) >ppk_1 1081 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -40.00) >psp_1 1080 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -37.80) >pss_1 1080 120 - 1 GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUUACCUAAUACGUGAUUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... ( -37.80) >consensus GCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAGCACUUUAAGUUGGGAGGAAGGGCAGUAAGUUAAUACCUUGCUGUUUUGACGUUACCGACAGAAUAAGCACCGGCUAACUC .....((((.(((((((.....((((....))))......))).....(((((..(..(((((((((((.......)))))))))))..)....))))).......))))))))...... (-41.53 = -39.07 + -2.46)

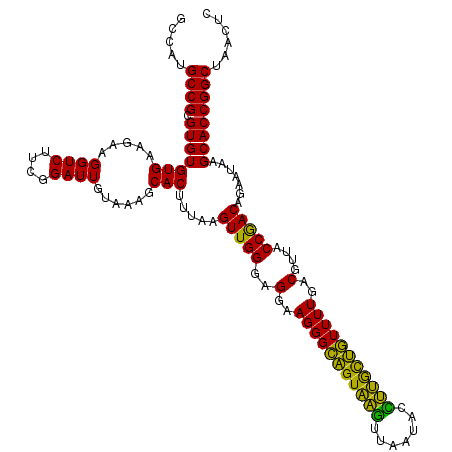
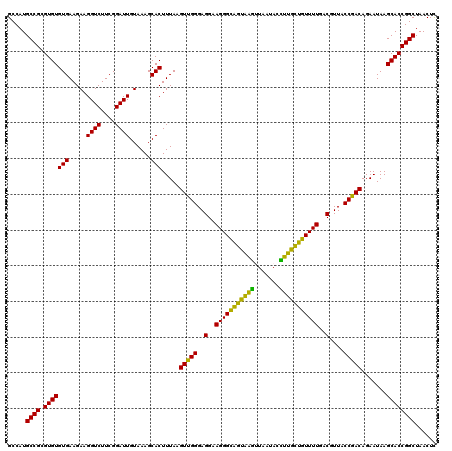
| Location | 4,793,306 – 4,793,426 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -33.70 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793306 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >pfo_1 1161 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >pfs_1 1161 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >ppk_1 1161 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >psp_1 1160 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >pss_1 1160 120 + 1 CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) ( -33.70) >consensus CUUUACAAUCCGAAGACCUUCUUCACACACGCGGCAUGGCUGGAUCAGGCUUUCGCCCAUUGUCCAAUAUUCCCCACUGCUGCCUCCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUG ...........((((.....))))...((((((((((((.(((((..(((....)))....))))).......))).))))))........((((.((((((...))).))))))).))) (-33.70 = -33.70 + -0.00)

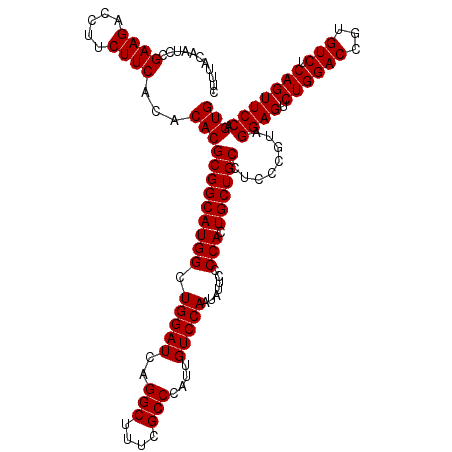
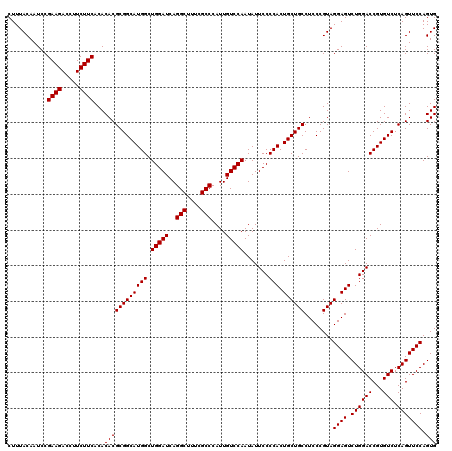
| Location | 4,793,306 – 4,793,426 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -42.50 |
| Energy contribution | -42.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793306 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >pfo_1 1161 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >pfs_1 1161 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >ppk_1 1161 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >psp_1 1160 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >pss_1 1160 120 - 1 CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... ( -42.50) >consensus CACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCAGCCAUGCCGCGUGUGUGAAGAAGGUCUUCGGAUUGUAAAG ..(((((.(((.....)))))))).((((....))))((.(((.(((......(((((...(((((....))))).))))))))))).))..((.(((((.....))))).))....... (-42.50 = -42.50 + 0.00)

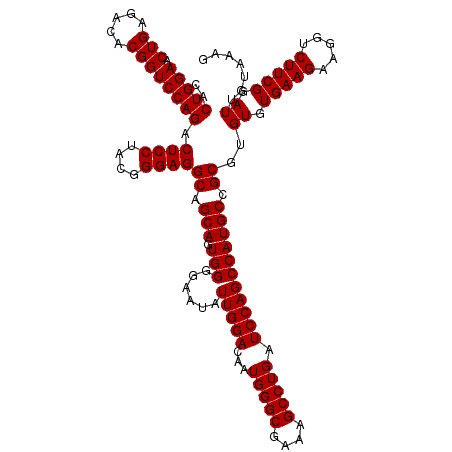
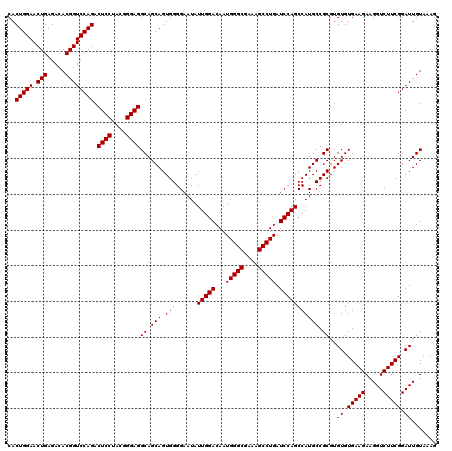
| Location | 4,793,346 – 4,793,466 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -42.50 |
| Energy contribution | -42.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793346 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >pfo_1 1201 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >pfs_1 1201 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >ppk_1 1201 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >psp_1 1200 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >pss_1 1200 120 - 1 AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) ( -42.50) >consensus AGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUGGGGAAUAUUGGACAAUGGGCGAAAGCCUGAUCCA .(....)..(((...(((((((.........)))))))(((.(((((.(((.....)))))))).((((....))))......))).)))....((((...(((((....))))).)))) (-42.50 = -42.50 + 0.00)

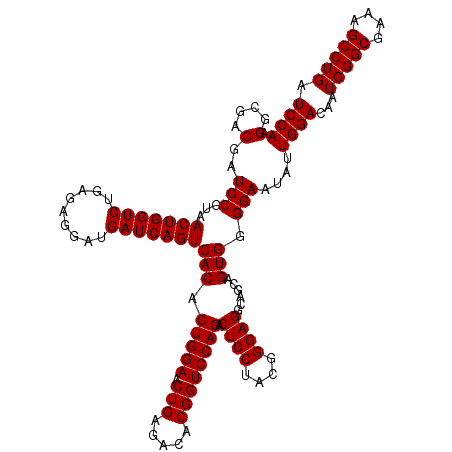
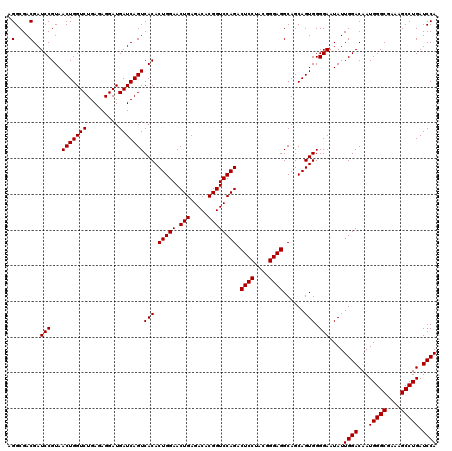
| Location | 4,793,386 – 4,793,506 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -46.77 |
| Consensus MFE | -47.29 |
| Energy contribution | -46.77 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.17 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793386 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -46.90) >pel_1 1241 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -46.20) >pfs_1 1241 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -47.10) >ppk_1 1241 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -46.20) >psp_1 1240 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -47.10) >pss_1 1240 120 - 1 CUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... ( -47.10) >consensus CUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCA .......(((((.(((..((((((((.......)))))))))))...)))))((.(((((((.........))))))).)).(((((.(((.....)))))))).((((....))))... (-47.29 = -46.77 + -0.53)

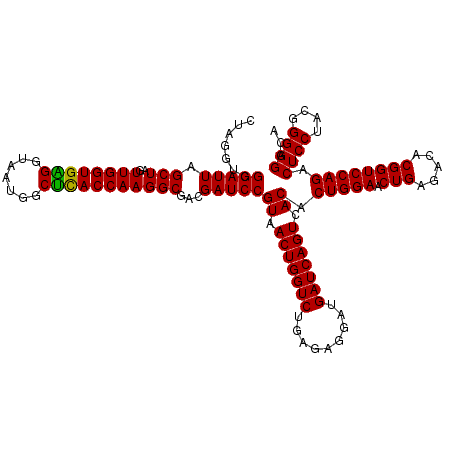

| Location | 4,793,426 – 4,793,546 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -30.94 |
| Consensus MFE | -28.93 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793426 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUAGGCCUUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGUGAGGUCCGAAGAUCCCCCACUUUC ((((((.((.........)).)))))).(((((...((.(((((.((.......)).)))))...))...((.((((((.(((........))).).))))).)).)))))......... ( -30.70) >pel_1 1281 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGCUUUC ((((((.((.........)).)))))).(((((.(((((((((((((((..............................))))))).........))))))..)).)))))......... ( -28.31) >pf5_1 1281 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUCCGACCUAGGCUCAUCUAAUAGCGUGAGGUCCGAAGAUCCCCCACUUUC ((((((.((.........)).)))))).(((((...((.((((((((.......))))))))...))...((.((((((.(((........))).).))))).)).)))))......... ( -35.10) >ppk_1 1281 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGCUUUC ((((((.((.........)).)))))).(((((.(((((((((((((((..............................))))))).........))))))..)).)))))......... ( -28.31) >psp_1 1280 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGCUUUC ((((((.((.........)).)))))).(((((.(((((((((((((.......)))))).....((((..((......)).((....))))))..)))))..)).)))))......... ( -31.60) >pss_1 1280 120 + 1 UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCUCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGCUUUC ((((((.((.........)).)))))).(((((.(((((((((((((.......)))))).....((((..((......)).((....))))))..)))))..)).)))))......... ( -31.60) >consensus UGACUGAUCAUCCUCUCAGACCAGUUACGGAUCGUCGCCUUGGUGAGCCAUUACCCCACCAACUAGCUAAUCCGACCUAGGCUCAUCUGAUAGCGCAAGGCCCGAAGGUCCCCUGCUUUC ((((((.((.........)).)))))).(((((.(((((((((((((.......)))))).....((((.((.((..........)).))))))..)))))..)).)))))......... (-28.93 = -28.77 + -0.17)
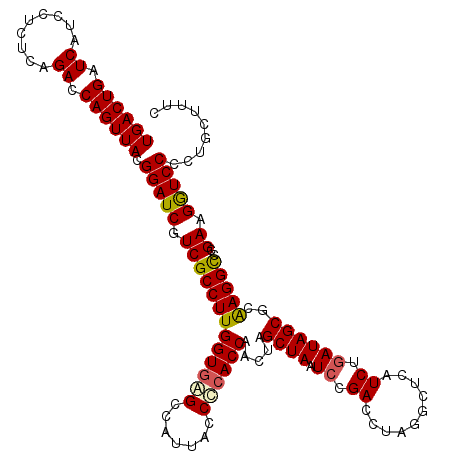
| Location | 4,793,426 – 4,793,546 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -44.78 |
| Consensus MFE | -46.13 |
| Energy contribution | -44.13 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.10 |
| Structure conservation index | 1.03 |
| SVM decision value | 5.01 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793426 120 - 1 GAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ((.((((((((.((....)))))).)))).)).(((((..((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....)))...))).)). ( -45.30) >pel_1 1281 120 - 1 GAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ....((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....))). ( -44.50) >pf5_1 1281 120 - 1 GAAAGUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ...((((((((.((....)))))).))))....(((((..((((..((((((.(((..((((((((.......)))))))))))...))))))...))))(((....)))...))).)). ( -43.60) >ppk_1 1281 120 - 1 GAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ....((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....))). ( -44.50) >psp_1 1280 120 - 1 GAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ....((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....))). ( -45.40) >pss_1 1280 120 - 1 GAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ....((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....))). ( -45.40) >consensus GAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCAAGGCGACGAUCCGUAACUGGUCUGAGAGGAUGAUCAGUCA ....((((((..((....)))))))).((.(((((.....((((..((((((.(((..((((((((.......)))))))))))...))))))...))))))))).))(((.....))). (-46.13 = -44.13 + -2.00)
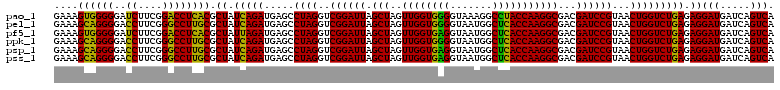
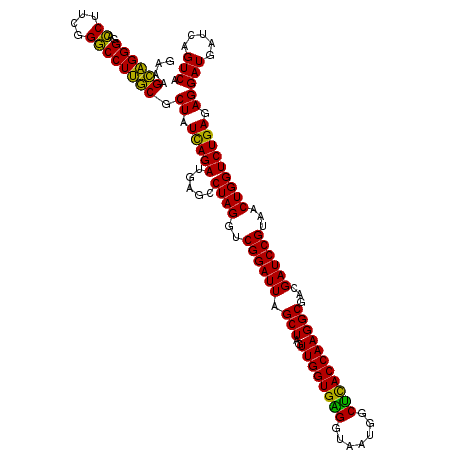
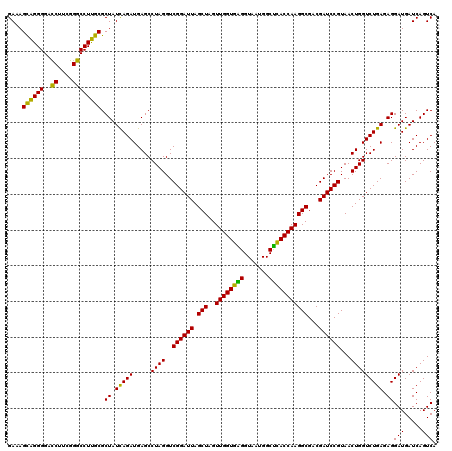
| Location | 4,793,466 – 4,793,586 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -47.63 |
| Consensus MFE | -49.90 |
| Energy contribution | -46.82 |
| Covariance contribution | -3.08 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.59 |
| Structure conservation index | 1.05 |
| SVM decision value | 5.78 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793466 120 - 1 GUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAAGGCCUACCA (((((....)))))((((((.(((....((((.(((...((...((((((..((....)))))))).)).))))))).((....)))))))))))....(((((((.......))))))) ( -49.40) >pel_1 1321 120 - 1 GUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCA (((((....)))))((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))....(((((((.......))))))) ( -46.60) >pf5_1 1321 120 - 1 GUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCA (((((....)))))((((((.((((.....(((....)))...((((((((.((....)))))).)))).((((......))))).)))))))))....(((((((.......))))))) ( -50.00) >pfo_1 1321 120 - 1 GUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCA (((((....)))))((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))....(((((((.......))))))) ( -47.50) >ppk_1 1321 120 - 1 GUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGGGGUAAUGGCUCACCA (((((....)))))((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))....(((((((.......))))))) ( -46.60) >pss_1 1320 120 - 1 GCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCA (..((....))..)((((((.((((.....(((....)))((.(((.((((.((....))))))..))).))............).)))))))))....(((((((.......))))))) ( -45.70) >consensus GUUCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGCCUAGGUCGGAUUAGCUAGUUGGUGAGGUAAUGGCUCACCA (((((....)))))((((((.((((.....(((....)))....((((((..((....))))))))(((........)))....).)))))))))....(((((((.......))))))) (-49.90 = -46.82 + -3.08)
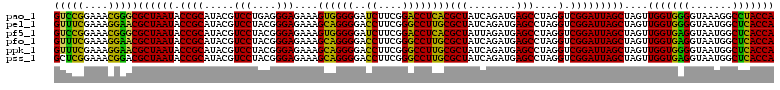
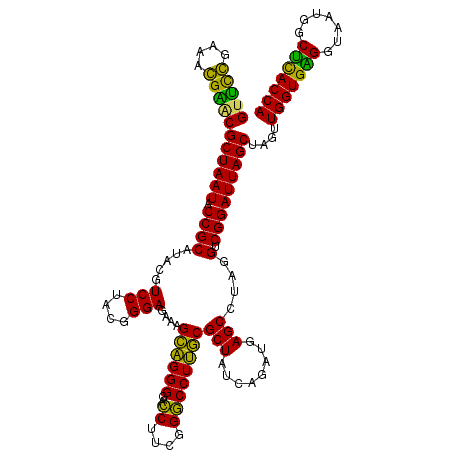
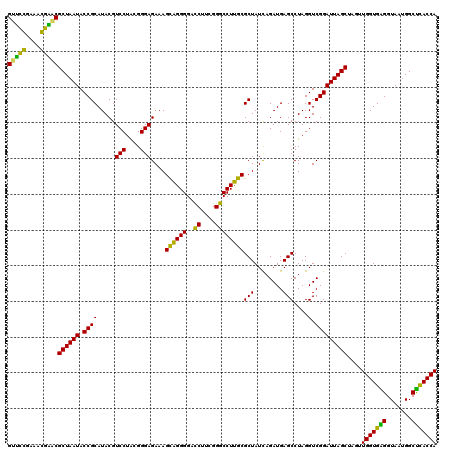
| Location | 4,793,506 – 4,793,626 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -43.04 |
| Consensus MFE | -43.56 |
| Energy contribution | -41.17 |
| Covariance contribution | -2.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793506 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUCAGAUGAGC .......((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).((((.(((...((...((((((..((....)))))))).)).)))))))... ( -45.00) >pel_1 1361 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGC ...(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))........ ( -42.82) >pf5_1 1361 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUAGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAAGUGGGGGAUCUUCGGACCUCACGCUAUUAGAUGAGC ...(((....(((((......)))))..((((....((.((((((....)))))).))...)))).))).(((....)))...((((((((.((....)))))).))))........... ( -43.70) >pfo_1 1361 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGC ...(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))........ ( -42.82) >ppk_1 1361 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGC ...(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))((.(((.((((.((....))))))..))).))........ ( -42.82) >pss_1 1360 120 - 1 UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGC ...(((.((((.(((......)))))))((((....((.((..((....))..)).))...)))).))).(((....)))((.(((.((((.((....))))))..))).))........ ( -41.10) >consensus UGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCGCUAUCAGAUGAGC ...(((.((((.(((......)))))))((((.......((((((....))))))......)))).))).(((....)))....((((((..((....))))))))(((........))) (-43.56 = -41.17 + -2.39)
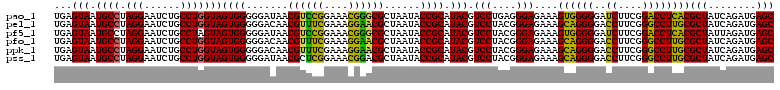
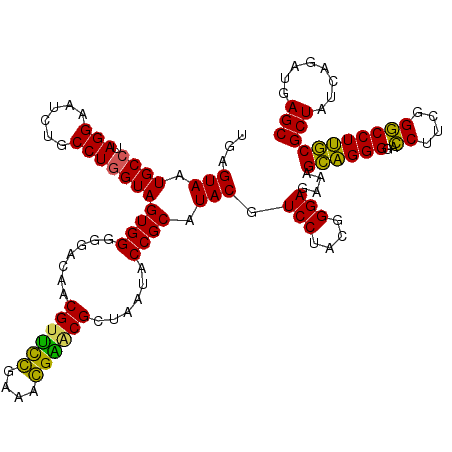
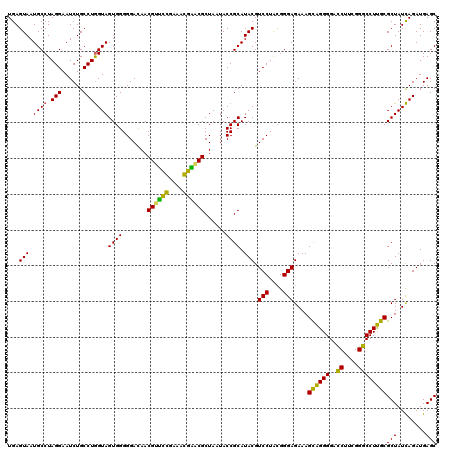
| Location | 4,793,519 – 4,793,639 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -46.72 |
| Consensus MFE | -48.46 |
| Energy contribution | -45.90 |
| Covariance contribution | -2.55 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4793519 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUGAGGGAGAAAGUGGGGGAUCUUCGGACCUCACG ......(((((.........((((.(((......)))))))(((.((..((.((((((....)))))).))...)).))).)))))...........((((((..((....)))))))). ( -46.60) >pel_1 1374 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCG .((((((((.((((.......)))).....)))))).((((...(....)..((((((....))))))....)))))).....(((....)))....((((((..((....)))))))). ( -46.90) >pf5_1 1374 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUAGUAGUGGGGGAUAACGUCCGGAAACGGGCGCUAAUACCGCAUACGUCCUACGGGAGAAAGUGGGGGAUCUUCGGACCUCACG ..((..(((((.(((.(((..((...(..((((.((.......)).))))..)(((((....)))))))...))))))...)))))..)).......((((((..((....)))))))). ( -48.50) >pfo_1 1374 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCG .((((((((.((((.......)))).....)))))).((((...(....)..((((((....))))))....)))))).....(((....)))....((((((..((....)))))))). ( -46.90) >ppk_1 1374 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUUCGAAAGGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCG .((((((((.((((.......)))).....)))))).((((...(....)..((((((....))))))....)))))).....(((....)))....((((((..((....)))))))). ( -46.90) >pss_1 1373 120 - 1 AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGAUAACGCUCGGAAACGGACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCG ..((..(((((.........((((.(((......)))))))(((.((..((.((..((....))..)).))...)).))).)))))..)).......((((((..((....)))))))). ( -44.50) >consensus AGCGGCGGACGGGUGAGUAAUGCCUAGGAAUCUGCCUGGUAGUGGGGGACAACGUUCCGAAACGAACGCUAAUACCGCAUACGUCCUACGGGAGAAAGCAGGGGACCUUCGGGCCUUGCG ...((((((.((((.......)))).....))))))..(((((((.......((((((....))))))......)))).))).(((....)))....((((((..((....)))))))). (-48.46 = -45.90 + -2.55)

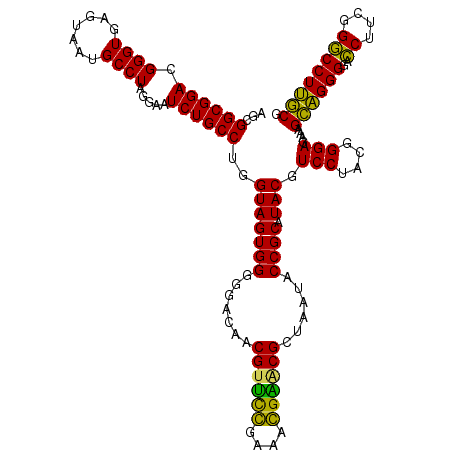
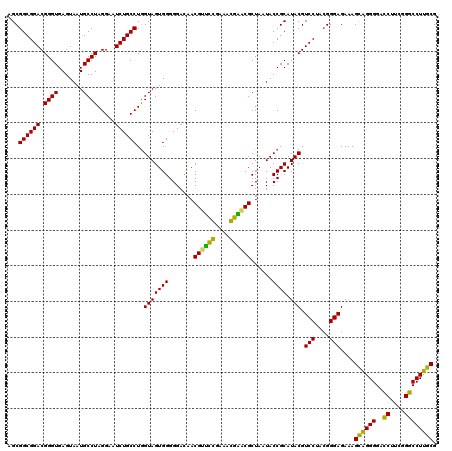
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:50:51 2007