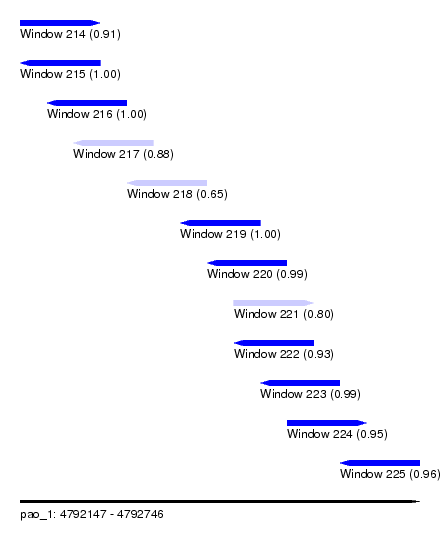
| Sequence ID | pao_1 |
|---|---|
| Location | 4,792,147 – 4,792,746 |
| Length | 599 |
| Max. P | 0.999993 |

| Location | 4,792,147 – 4,792,267 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -36.04 |
| Energy contribution | -36.32 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
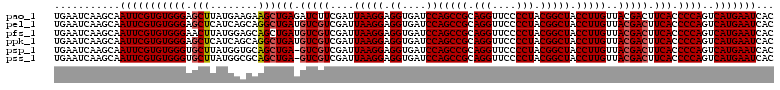
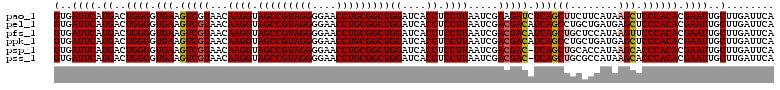
>pao_1 4792147 120 + 1 UGAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAGAUCUUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........((((((((((((((((......)))))..((...(((..(((((.((....))(((((.(((....))).))))).)))))...)))..))..))))..)))))))... ( -36.30) >pel_1 1 120 + 1 UGAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGCAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........(((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))))... ( -41.70) >pfs_1 1 120 + 1 UGAAUCAAGCAAUUCGUGUGGGAACUUAUGGAGCAGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC (((.((((((..((((((.(....).))))))...)))((.(((((....(((((.((....))(((((.(((....))).))))).)))))..))))).))..........))).))). ( -36.50) >ppk_1 1 120 + 1 UGAAUCAAGCAAUUCGUGUGGGAGCUCAUCAGCAGGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........(((((((((((....((((((....))))))..((((..(((((.((....))(((((.(((....))).))))).)))))...)))).....))))..)))))))... ( -41.70) >psp_1 1 119 + 1 UGAAUCAAGCAAUUCGUGUGGGUGCUUAUGGUGCAGCUGA-GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........((((((((((((((.......)))...((-(((((....(((((.((....))(((((.(((....))).))))).)))))..)))))))...))))..)))))))... ( -38.50) >pss_1 1 119 + 1 UGAAUCAAGCAAUUCGUGUGGGUGCUUAUGGCGCAGCUGA-GUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........(((((((.(((((.....(((...)))((-(((((....(((((.((....))(((((.(((....))).))))).)))))..))))))))))))....)))))))... ( -39.10) >consensus UGAAUCAAGCAAUUCGUGUGGGAGCUUAUGAAGAAGCUGAUGUCGUCGAUUAAGGAGGUGAUCCAGCCGCAGGUUCCCCUACGGCUACCUUGUUACGACUUCACCCCAGUCAUGAAUCAC ...........(((((((((((.(((........)))(((.(((((....(((((.((....))(((((.(((....))).))))).)))))..))))).))).))))..)))))))... (-36.04 = -36.32 + 0.28)
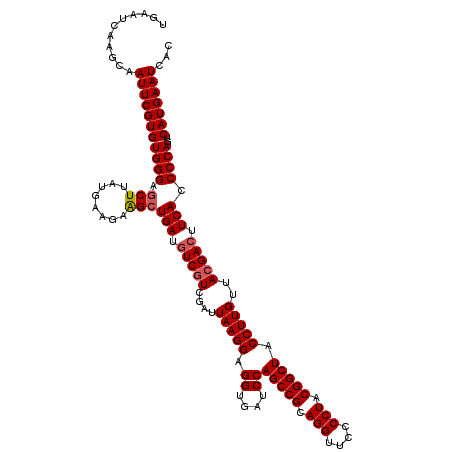
| Location | 4,792,147 – 4,792,267 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.61 |
| Mean single sequence MFE | -46.25 |
| Consensus MFE | -43.43 |
| Energy contribution | -43.65 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792147 120 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((...(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........)))))...........)).))))..)........ ( -41.80) >pel_1 1 120 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((..((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).)))))).))))..)........ ( -49.40) >pfs_1 1 120 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCUCCAUAAGUUCCCACACGAAUUGCUUGAUUCA (..((((.((..((((((((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........)))))))))).))))..)........ ( -44.10) >ppk_1 1 120 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCCUGCUGAUGAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((..((((((...((((....((((.(((((((((....)))))))))((....)).))))...))))..((((((....)))))).)).)))))).))))..)........ ( -49.40) >psp_1 1 119 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCACCAUAAGCACCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).)))))).))))..)........ ( -46.40) >pss_1 1 119 - 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCGCCAUAAGCACCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).)))))).))))..)........ ( -46.40) >consensus GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCCCCAUAAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))).)))))).))))..)........ (-43.43 = -43.65 + 0.22)
| Location | 4,792,187 – 4,792,307 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -49.63 |
| Consensus MFE | -48.85 |
| Energy contribution | -48.85 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.72 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
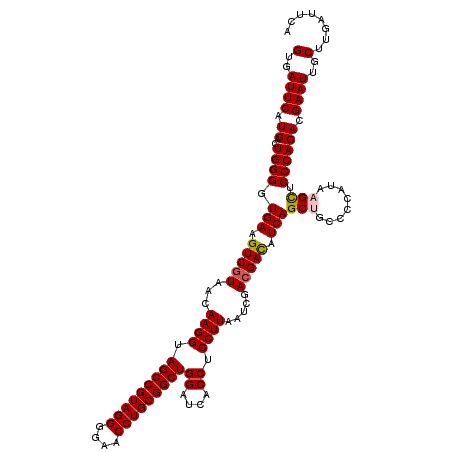
>pao_1 4792187 120 - 1 AAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUC ..((((((.((((..((....)).))))))))))...((((((((((((((((.......))))))........(((((((((....))))))))))))))).))))............. ( -51.90) >pel_1 41 120 - 1 AAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACA ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............. ( -49.60) >pfs_1 41 120 - 1 AAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACA ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............. ( -48.90) >ppk_1 41 120 - 1 AAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACA ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............. ( -48.90) >psp_1 41 119 - 1 AAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC- ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............- ( -49.60) >pss_1 41 119 - 1 AAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC- ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............- ( -48.90) >consensus AAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACA ..((((((.((((..((....)).))))))))))...((.(((((((((((((.......))))))........(((((((((....))))))))))))))))..))............. (-48.85 = -48.85 + 0.00)

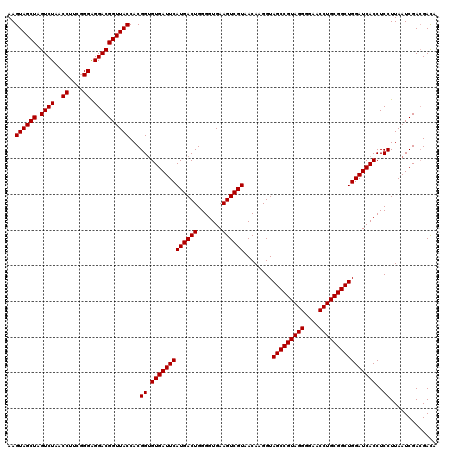
| Location | 4,792,227 – 4,792,347 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -42.03 |
| Energy contribution | -41.75 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
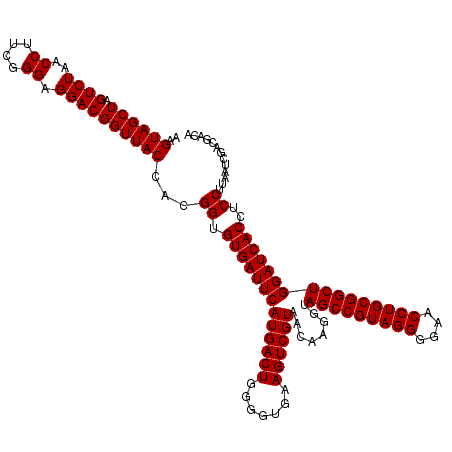
>pao_1 4792227 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU ....(((..(((((......))))).)))(((((((.....))))))).((((..((....)).))))(((((((..(((....)))((((((.......)))))).....))))))).. ( -43.40) >pel_1 81 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... ( -42.00) >pfs_1 81 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... ( -41.30) >ppk_1 81 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... ( -41.30) >psp_1 80 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... ( -42.00) >pss_1 80 120 - 1 UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... ( -41.30) >consensus UACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGU .....(((...((.(.((((..((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..))..))))..).))......)))...... (-42.03 = -41.75 + -0.28)


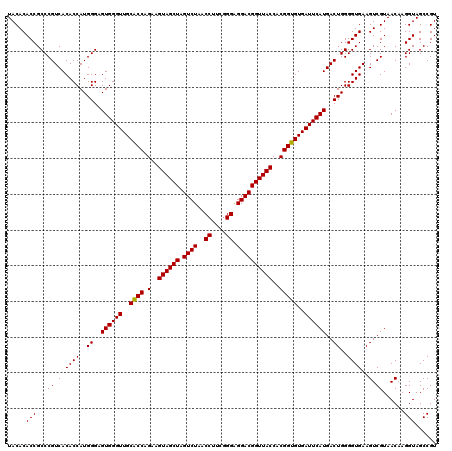
| Location | 4,792,307 – 4,792,427 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -40.72 |
| Energy contribution | -40.17 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.69 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792307 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGUGAAUCAGAAUGUCACGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAG ...(((((((.(((.(((((..((.((.((((.(....).)))).)).))...))))((((..........(((((..((....))..)))))...))))...).))))))))))..... ( -38.92) >pel_1 161 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCAAAUCAGAAUGUUGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ..(((((((((..(.((((.....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..)))))......)))).)..))))))))).... ( -39.50) >pfs_1 161 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ..(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..)))))))))))))).... ( -41.90) >ppk_1 161 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ..(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..)))))))))))))).... ( -41.90) >psp_1 160 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ..(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..)))))))))))))).... ( -41.90) >pss_1 160 120 - 1 UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ..(((((((((((((((.....((.((.((((.(....).)))).)).))....)))))).......(((((((((..((....))..))))(......)..)))))))))))))).... ( -41.90) >consensus UCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUCGCGAAUCAGAAUGUCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAG ...((((((((..(.((((.....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..)))))......)))).)..))))))))..... (-40.72 = -40.17 + -0.55)
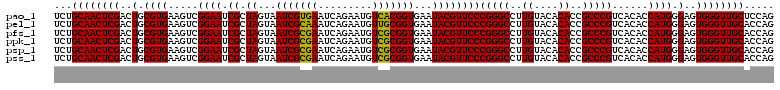
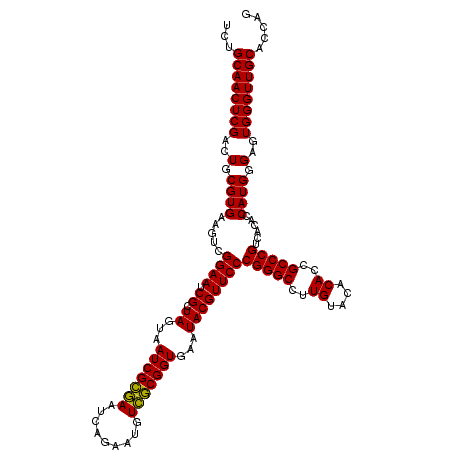
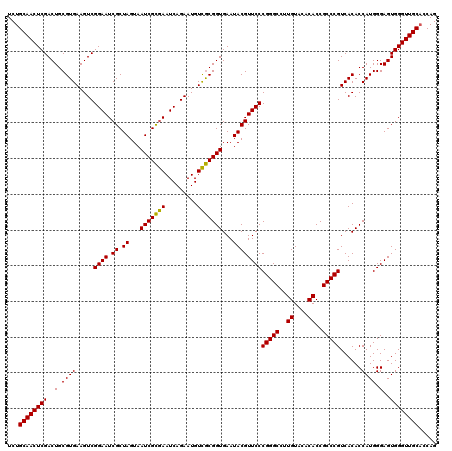
| Location | 4,792,387 – 4,792,507 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.56 |
| Mean single sequence MFE | -44.58 |
| Consensus MFE | -44.19 |
| Energy contribution | -43.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.36 |
| SVM RNA-class probability | 0.999879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792387 120 - 1 GCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.80) >pf5_1 241 120 - 1 GCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.80) >pfo_1 241 120 - 1 GCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((.....(((..((...(((....)))...))....))).....)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.80) >pfs_1 241 120 - 1 GCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((...(.(((..((...(((....)))...))....))).)...)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.90) >psp_1 240 120 - 1 GCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((...((((...((...(((....)))...))...)))).....)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.10) >pss_1 240 120 - 1 GCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((...((((...((...(((....)))...))...)))).....)))))))))..(((((..(((((((........))))))).....)))))...).)))).... ( -44.10) >consensus GCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGUCGGAAUCGCUAGUAAUC (((((.(((((((((.....(((((((...(((....)))...)).)))))......)))))))))..(((((..(((((((........))))))).....)))))...).)))).... (-44.19 = -43.97 + -0.22)

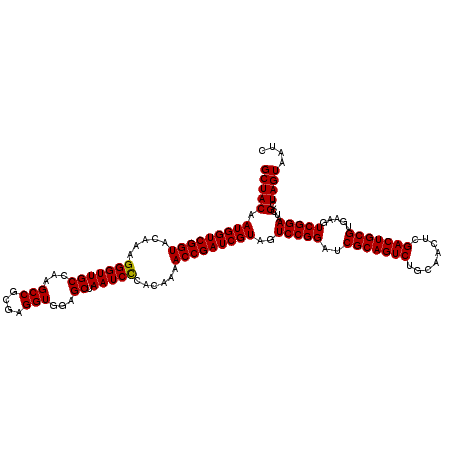
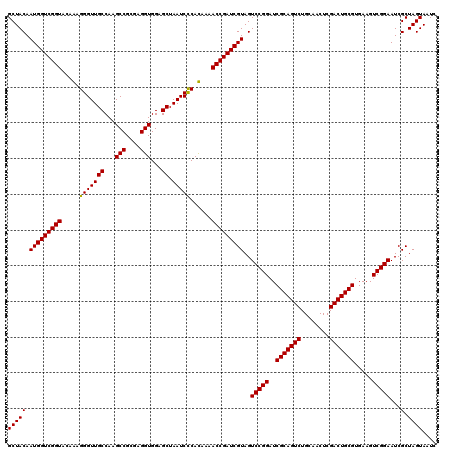
| Location | 4,792,427 – 4,792,547 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.28 |
| Mean single sequence MFE | -43.65 |
| Consensus MFE | -43.01 |
| Energy contribution | -42.78 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792427 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((.(((((.....)))))(((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))).))).)).. ( -45.20) >pf5_1 281 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....))))((((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))))))).)).. ( -43.60) >pfo_1 281 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCAUAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....))))((((((((....(.....)...(((((((.....(((..((...(((....)))...))....))).....)))))))))))))))))).)).. ( -43.60) >pfs_1 281 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....))))((((((((....(.....)...(((((((...(.(((..((...(((....)))...))....))).)...)))))))))))))))))).)).. ( -43.70) >psp_1 280 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....))))((((((((....(.....)...(((((((...((((...((...(((....)))...))...)))).....)))))))))))))))))).)).. ( -42.90) >pss_1 280 120 - 1 GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCUCACAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....))))((((((((....(.....)...(((((((...((((...((...(((....)))...))...)))).....)))))))))))))))))).)).. ( -42.90) >consensus GUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGUGGAGCUAAUCCCACAAAACCGAUCGUAGUCCGGAUCGCAG .....((.(((..((((.....)))).(((((((....(.....)...(((((((.....(((((((...(((....)))...)).)))))......)))))))))))))).))).)).. (-43.01 = -42.78 + -0.22)


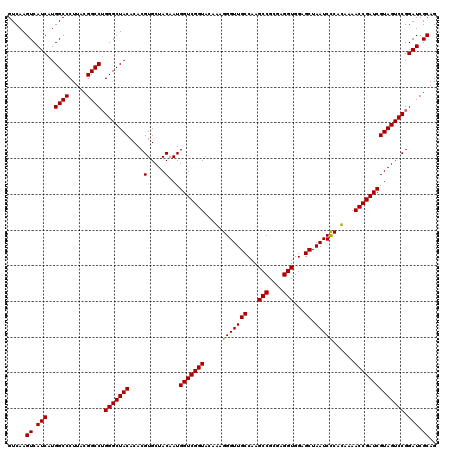
| Location | 4,792,467 – 4,792,587 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.22 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -37.38 |
| Energy contribution | -37.55 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792467 120 + 1 ACCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCUGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((.(((.((.(..(((..(((...)))..)))...).)).)))..)))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -37.00) >pf5_1 321 120 + 1 ACCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(..(((..(((...)))..)))...).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -38.00) >pfo_1 321 120 + 1 ACCUCGCGGCUUGGCAACCCUUUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(..(((..(((...)))..)))...).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -38.00) >ppk_1 321 120 + 1 ACCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -38.90) >psp_1 320 120 + 1 ACCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -38.90) >pss_1 320 120 + 1 ACCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(.((((..(((...)))..))).).).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... ( -38.90) >consensus ACCUCGCGGCUUGGCAACCCUCUGUACCGACCAUUGUAGCACGUGUGUAGCCCAGGCCGUAAGGGCCAUGAUGACUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCC .(((..(((((((((.((.(..(((..(((...)))..)))...).)).).))))))))..)))(((..((((((.....)))))).............(((.....))))))....... (-37.38 = -37.55 + 0.17)

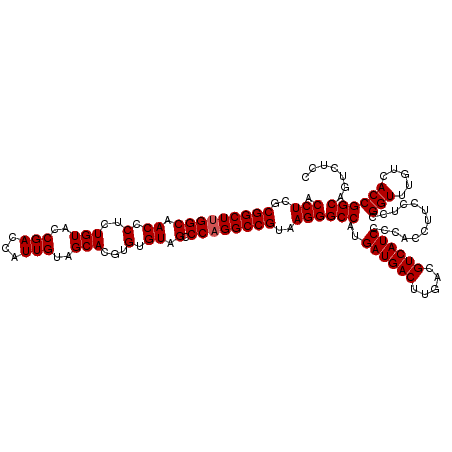
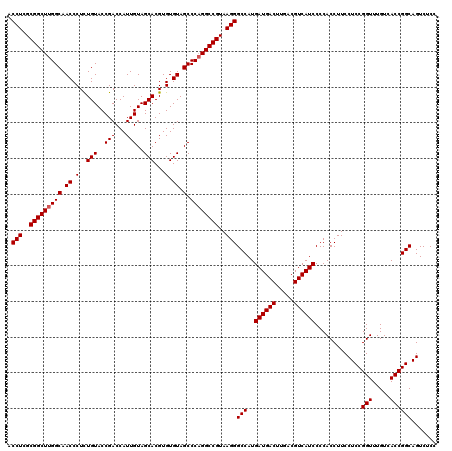
| Location | 4,792,467 – 4,792,587 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.22 |
| Mean single sequence MFE | -45.27 |
| Consensus MFE | -45.03 |
| Energy contribution | -44.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792467 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.((((((.(((.((....)))))....)))))).....))))))))))..(((....))) ( -46.90) >pf5_1 321 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) ( -44.70) >pfo_1 321 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) ( -44.70) >ppk_1 321 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) ( -45.10) >psp_1 320 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) ( -45.10) >pss_1 320 120 - 1 GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAGAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) ( -45.10) >consensus GGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGUGCUACAAUGGUCGGUACAAAGGGUUGCCAAGCCGCGAGGU .....((.(((((.....))))).))..(((...((((((.....))))))..(((((((.(((((..(((.((....))))).....))))).....))))))))))..(((....))) (-45.03 = -44.78 + -0.25)
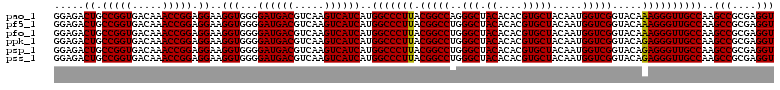
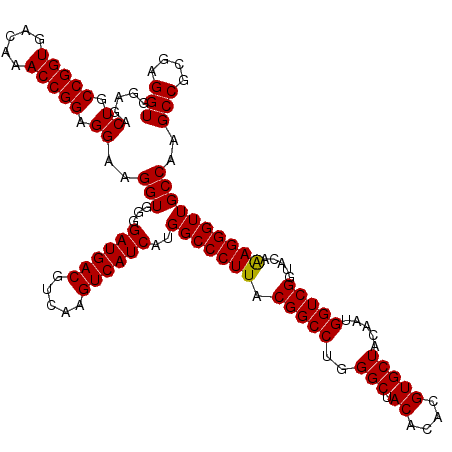

| Location | 4,792,507 – 4,792,626 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -45.25 |
| Consensus MFE | -44.12 |
| Energy contribution | -44.28 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792507 119 - 1 CUUGUCCUUAGUUACCAGCACCUCG-GGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCAGGGCUACACACGU ...((((.......(((.(.((((.-(((.((((.(((....))).))))(....)..))).))))..).))).((((((.....)))))).(((((.....)))))))))......... ( -48.00) >pfo_1 361 120 - 1 CUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... ( -44.70) >pfs_1 361 120 - 1 CUUGUCCUUAGUUACCAGCACGUCAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... ( -44.70) >ppk_1 361 120 - 1 CUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... ( -44.70) >psp_1 360 120 - 1 CUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... ( -44.70) >pss_1 360 120 - 1 CUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... ( -44.70) >consensus CUUGUCCUUAGUUACCAGCACGUUAUGGUGGGCACUCUAAGGAGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAGUCAUCAUGGCCCUUACGGCCUGGGCUACACACGU (((.(((((((...(((.((.....)).))).....)))))))..((.(((((.....))))).)))))(((..((((((.....)))))).((((((.........))))))))).... (-44.12 = -44.28 + 0.17)

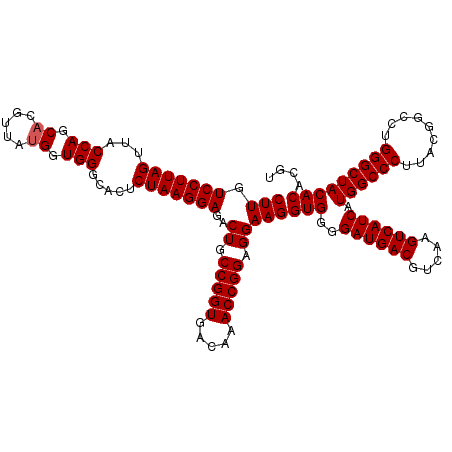
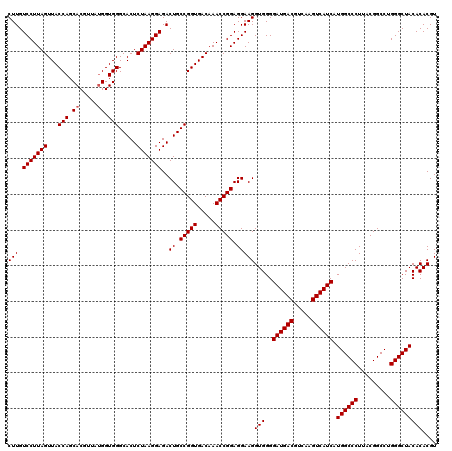
| Location | 4,792,547 – 4,792,666 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -34.23 |
| Energy contribution | -34.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792547 119 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACC-CGAGGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..((((((((-...))))..)))).)))))))...((((((.(((((...)))).).))))))..........))) ( -36.00) >pfo_1 401 120 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))) ( -33.90) >pfs_1 401 120 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUGACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((.......((((...(((((.(((((..((((.(((....))).)))).....))))).))))).....))))....((((((.(((((...)))).).))))))..........))) ( -37.40) >ppk_1 401 120 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))) ( -33.90) >psp_1 400 120 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))) ( -33.90) >pss_1 400 120 + 1 GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))) ( -33.90) >consensus GUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCUCCUUAGAGUGCCCACCAUAACGUGCUGGUAACUAAGGACAAGGGUUGCGCUCGUUACGGGACUUAACCCAACAUCUCACGAC (((............(((((((.....))))).))..(((((((..(((((((......)))..)))).)))))))...((((((.(((((...)))).).))))))..........))) (-34.23 = -34.23 + -0.00)

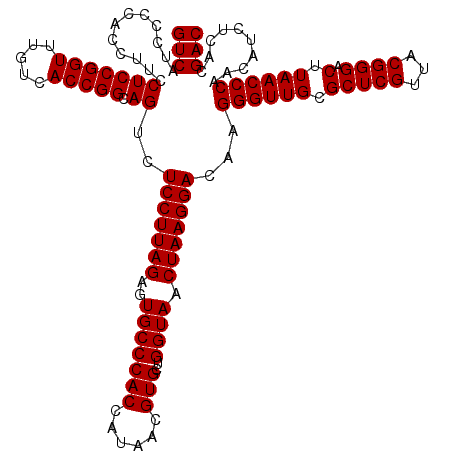
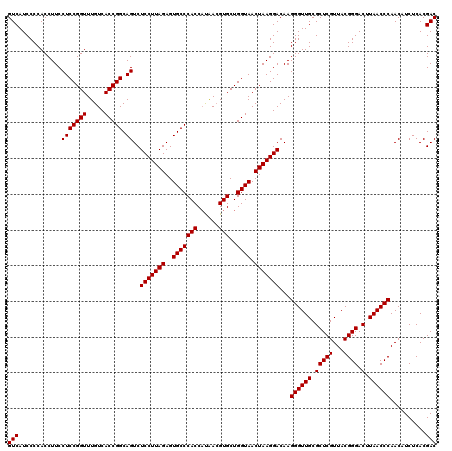
| Location | 4,792,626 – 4,792,746 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -43.05 |
| Energy contribution | -40.83 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.84 |
| Structure conservation index | 1.05 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4792626 120 - 1 ACAUGCUGAGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAACUCAGACACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -42.70) >pel_1 481 120 - 1 ACAUGCAGAGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAACUCUGACACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -42.70) >pf5_1 481 120 - 1 ACAUCCAAUGAACUUUCUAGAGAUAGAUUGGUGCCUUCGGGAACAUUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -38.40) >pfo_1 481 120 - 1 ACAUCCAAUGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAGCAUUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -40.10) >pfs_1 481 120 - 1 ACAUCCAAUGAACUUUCUAGAGAUAGAUUGGUGCCUUCGGGAACAUUGAGACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -38.40) >ppk_1 481 120 - 1 ACAUGCAGAGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAACUCUGACACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) ( -42.70) >consensus ACAUCCAAAGAACUUUCCAGAGAUGGAUUGGUGCCUUCGGGAACACUGACACAGGUGCUGCAUGGCUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGUAACGAGCGCAACC ...(((((((.(((.((((....))))..))).((....))..))))).))..(((..(((.((((....))))(((((((((....)))...(((......)))...)))))))))))) (-43.05 = -40.83 + -2.22)

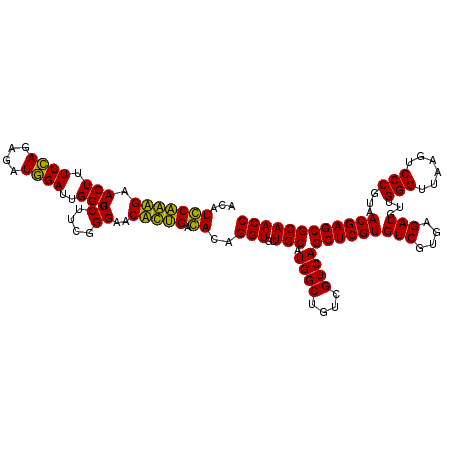
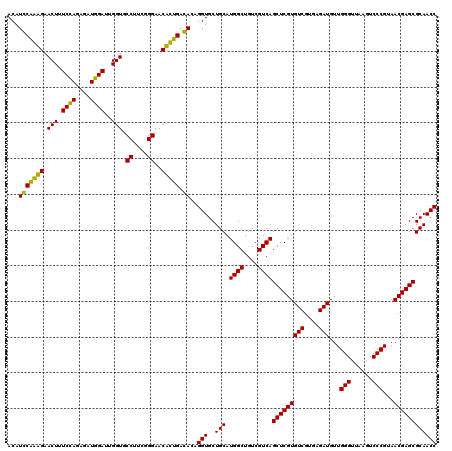
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:50:26 2007