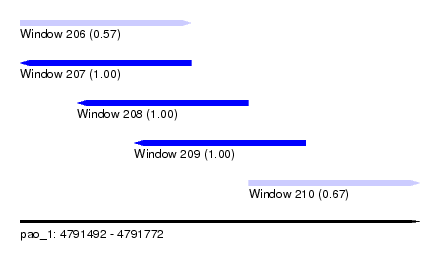
| Sequence ID | pao_1 |
|---|---|
| Location | 4,791,492 – 4,791,772 |
| Length | 280 |
| Max. P | 0.999999 |

| Location | 4,791,492 – 4,791,612 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -28.42 |
| Consensus MFE | -26.17 |
| Energy contribution | -24.12 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
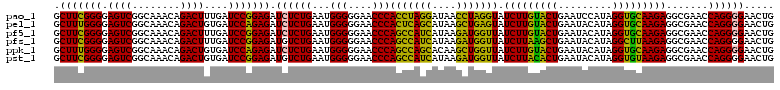
>pao_1 4791492 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCAUUCAG ((((.(..(((.(((......((((((.(((..((....)))))..))))))...((.....))......))).)))...).))))(((((((....)))))))(((....)))...... ( -29.30) >pel_1 2679 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUACUCAGCUUAUGCUGAGUGGGUUCCCCCAUUCAG .....((((((..((((...((..((((((.(((..(..((((.....))))..)((.....))..))).))))))))...))))...))))))....(((((((((....))))))))) ( -30.40) >pfs_1 2678 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUAAGCCUAUGUAUUCAGCUUAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAG .....(((((...........((((((.(((..((....)))))..))))))......((((((((...........))))))))))))).........((..((((....))))..)). ( -28.70) >ppk_1 2679 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUGUGCUGGCUGGGUUCCCCCAUUCAG .....(((.............((((((.(((..((....)))))..))))))...((.....))))).......((((((((((........)))))))))).((((....))))..... ( -28.50) >pss_1 2677 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAG ((((.(((.............((((((.(((..((....)))))..))))))...((.....)))))..(((((...)))))))))..(((((....))))).((((....))))..... ( -24.80) >pst_1 2677 120 + 1 AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUACACCUAUGUAUUCAGUGUAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAG .....(((((...........((((((.(((..((....)))))..))))))......((((((((...........))))))))))))).........((..((((....))))..)). ( -28.80) >consensus AUCUCGGUUGAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGAUGGCUGGGUUCCCCCAUUCAG .....(((.(((..........(((((.(((..((....)))))..)))))))).)))(((((.((...........)).)))))...(((((....))))).((((....))))..... (-26.17 = -24.12 + -2.05)
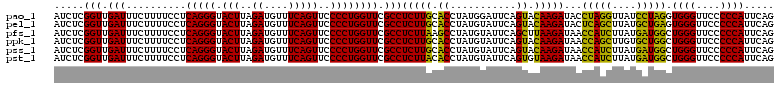
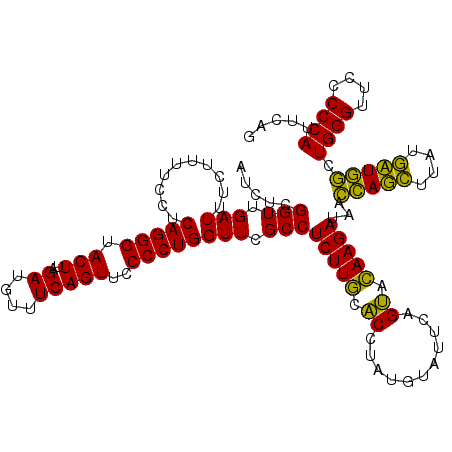
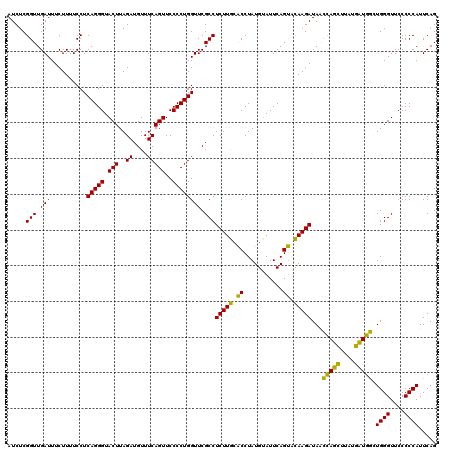
| Location | 4,791,492 – 4,791,612 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.67 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -39.50 |
| Energy contribution | -36.62 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.17 |
| Structure conservation index | 1.08 |
| SVM decision value | 6.85 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4791492 120 - 1 CUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -37.50) >pel_1 2679 120 - 1 CUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -37.90) >pfs_1 2678 120 - 1 CUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUAAGCUGAAUACAUAGGCUUAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -35.50) >ppk_1 2679 120 - 1 CUGAAUGGGGGAACCCAGCCAGCACAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -37.60) >pss_1 2677 120 - 1 CUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -35.60) >pst_1 2677 120 - 1 CUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... ( -35.60) >consensus CUGAAUGGGGGAACCCAGCCAGCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUCAACCGAGAU .....(((..((.((((((((((....))))))).(((((((((.........))))))))))).)..((.((((.(((..........))).))))..))........))..))).... (-39.50 = -36.62 + -2.88)

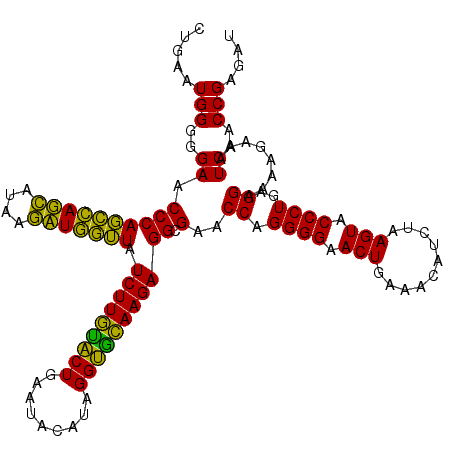
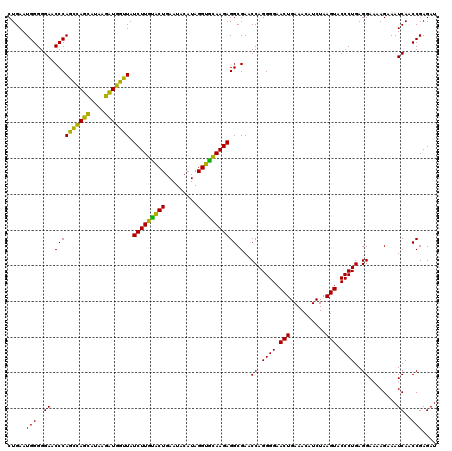
| Location | 4,791,532 – 4,791,652 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.83 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -47.11 |
| Energy contribution | -44.12 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.22 |
| Structure conservation index | 1.05 |
| SVM decision value | 6.33 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4791532 120 - 1 GCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUG .((((((((((((........)))))...))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))..... ( -47.30) >pel_1 2719 120 - 1 GCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUG .(((((((.((((........))))....))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))..... ( -45.80) >pf5_1 2715 120 - 1 GCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUG .((((((((((((........)))))...))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))..... ( -45.40) >pfs_1 2718 120 - 1 GCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUAAGCUGAAUACAUAGGCUUAAGAGGCGAACCAGGGGAACUG .((((((((((((........)))))...))))))).((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))...(((.....))) ( -42.70) >ppk_1 2719 120 - 1 GCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCACAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUG .(((((((.((((........))))....))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))..... ( -45.50) >pst_1 2717 120 - 1 GCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAGAGGCGAACCAGGGGAACUG .(((((((.((((........))))....))))))).((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))...(((.....))) ( -42.10) >consensus GCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUG .(((((((.((((........))))....))))))).((((((...(((....)))(((((((....))))))).(((((((((.........))))))))).......))))))..... (-47.11 = -44.12 + -2.99)
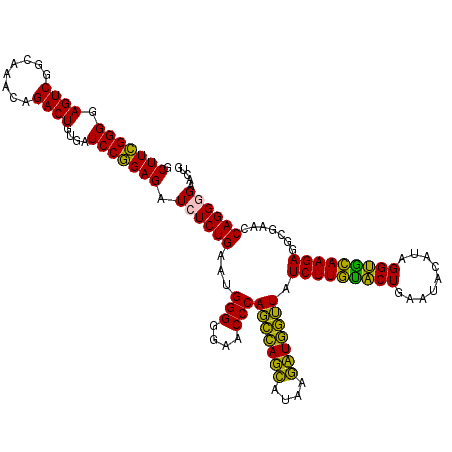
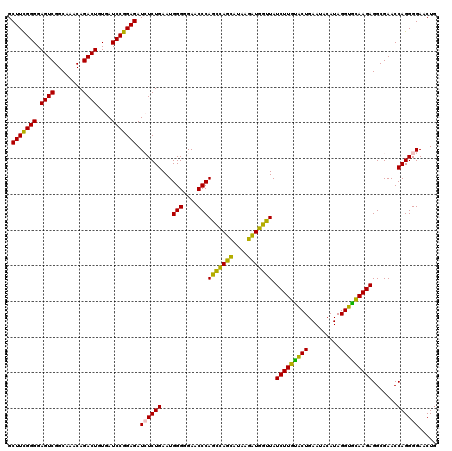
| Location | 4,791,572 – 4,791,692 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.39 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -42.02 |
| Energy contribution | -40.13 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.03 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4791572 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUG .....(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).)))))))..(((....)))(((((((....)))))))...... ( -43.30) >pel_1 2759 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUG .....(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))...... ( -41.80) >pf5_1 2755 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUG (((..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).))))))).((((....)))))))....((((((....)))))) ( -41.60) >pfs_1 2758 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUA (((..(((((.(..........((..(....)..)).....((((((((((((........)))))...))))))).).))))).((((....)))))))....((((((....)))))) ( -38.70) >ppk_1 2759 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCACAAGCUGGUUAUCUUG .....(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))...... ( -41.50) >pst_1 2757 120 - 1 GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUA (((..(((((.(..........((..(....)..)).....(((((((.((((........))))....))))))).).))))).((((....)))))))....((((((....)))))) ( -38.00) >consensus GGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGAUGGUUAUCUUG .....(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).)))))))..(((....)))(((((((....)))))))...... (-42.02 = -40.13 + -1.88)

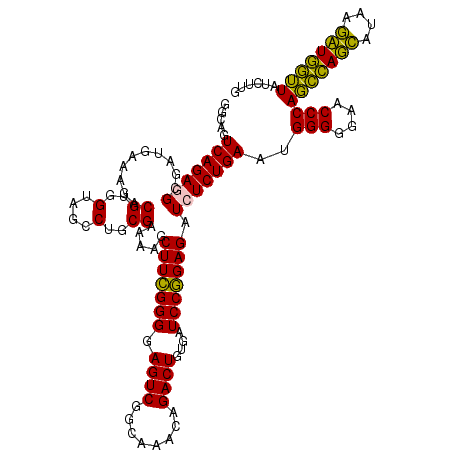
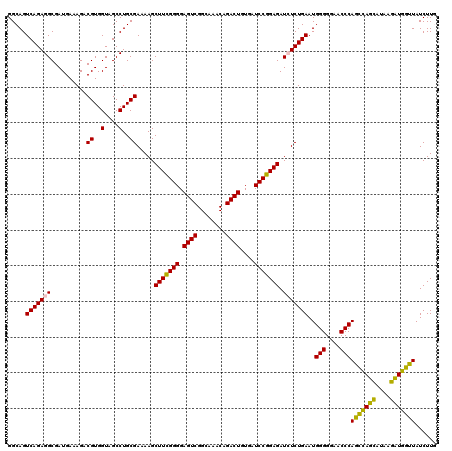
| Location | 4,791,652 – 4,791,772 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.03 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4791652 120 + 1 UUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCC ........(((................((((.((.....)))))).......(.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).)))) ( -25.20) >pel_1 2839 120 + 1 UUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCC ........(((................((((.((.....)))))).......(.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).)))) ( -25.20) >pf5_1 2835 120 + 1 UUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUAUGAAGACGACAUUCGCC ........(((.....((((((.(((.((((.((.....)))))).......(((((.(((......((((............))))......)))))))).)))))))))......))) ( -22.30) >pfs_1 2838 120 + 1 UUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACAACAUUCGCC ....(((((((......))).......((((.((.....))))))..........))))..(((((((.((((((.((...........)).))))))....)))))))........... ( -24.60) >ppk_1 2839 120 + 1 UUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCC ........(((................((((.((.....)))))).......(.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).)))) ( -25.20) >pst_1 2837 120 + 1 UUUUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCGAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCC ........(((................(((((........))))).......(.(((((..(((((((.((((((.((...........)).))))))....))))))))).))).)))) ( -28.30) >consensus UUAUCGCAGGCUACCACGUCUUUCAUCGCCUCUGACUGCCAAGGCAUCCACCGUAUGCGCUUCUUCACUUGACCAUAUAACCCCAAGCAAUCUGGUUAUACUGUGAAGACGACAUUCGCC ....(((((((......))).......((((.((.....))))))..........))))..(((((((.((((((.((...........)).))))))....)))))))........... (-24.00 = -24.03 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:50:06 2007