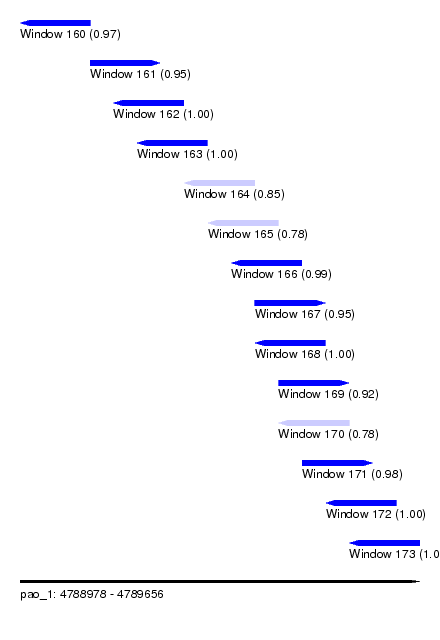
| Sequence ID | pao_1 |
|---|---|
| Location | 4,788,978 – 4,789,656 |
| Length | 678 |
| Max. P | 0.999338 |

| Location | 4,788,978 – 4,789,097 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -45.93 |
| Consensus MFE | -44.22 |
| Energy contribution | -44.38 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
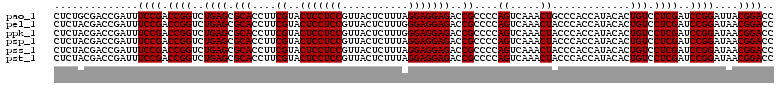
>pao_1 4788978 119 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGG-GUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((((........))))((((((.(......)))))))-...(((((((((((....))))))..))))))))))).................(((((...........))))). ( -43.90) >pel_1 161 120 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...(((....)))....(((((...........))))). ( -48.20) >pfo_1 161 120 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..))))))))))).................(((((...........))))). ( -45.10) >pfs_1 161 120 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAUAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..))))))))))).................(((((...........))))). ( -45.10) >ppk_1 161 120 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...(((....)))....(((((...........))))). ( -48.20) >pst_1 158 120 - 1 GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..))))))))))).................(((((...........))))). ( -45.10) >consensus GGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGG ((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..))))))))))).................(((((...........))))). (-44.22 = -44.38 + 0.17)

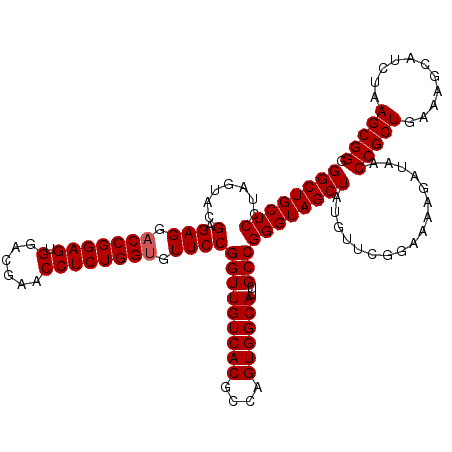
| Location | 4,789,097 – 4,789,216 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.72 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -29.67 |
| Energy contribution | -29.83 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789097 119 + 1 CCUCUCAAAUCUCAAACGUCCACGG-AGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .................((((((((-((...(((...((((.(((....)))))))....)))..))).)))..........(((((.....))))).......)))).(((....))). ( -33.30) >pfs_1 281 120 + 1 CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...))). ( -30.70) >ppk_1 281 120 + 1 CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...))). ( -30.70) >psp_1 278 120 + 1 CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...))). ( -30.70) >pss_1 278 120 + 1 CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...))). ( -30.70) >pst_1 278 120 + 1 CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...))). ( -30.70) >consensus CCUCUCAAAUCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCC .......................(((.((..(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).))))))))).))). (-29.67 = -29.83 + 0.17)

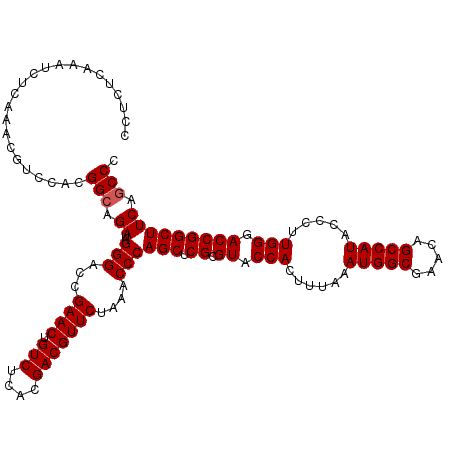
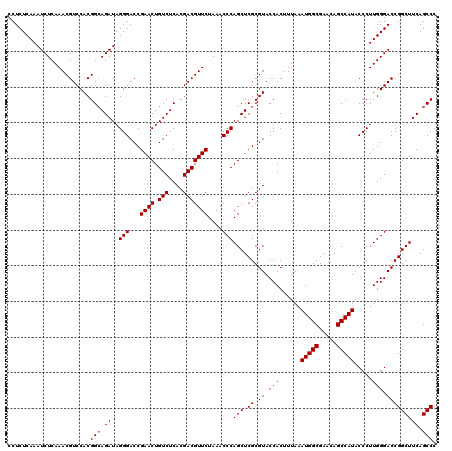
| Location | 4,789,136 – 4,789,256 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -48.50 |
| Consensus MFE | -48.50 |
| Energy contribution | -48.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789136 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >pfs_1 321 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >ppk_1 321 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >psp_1 318 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >pss_1 318 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >pst_1 318 120 - 1 GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. ( -48.50) >consensus GGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAG .(.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))).........(((....))).. (-48.50 = -48.50 + 0.00)

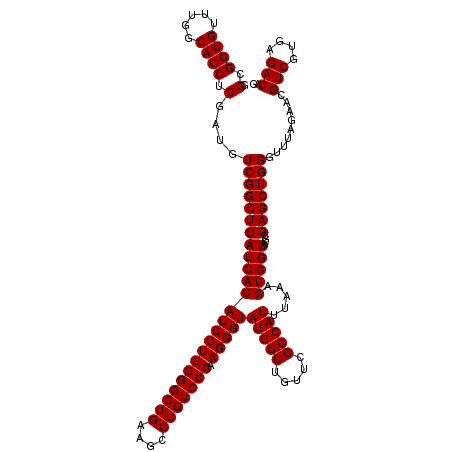
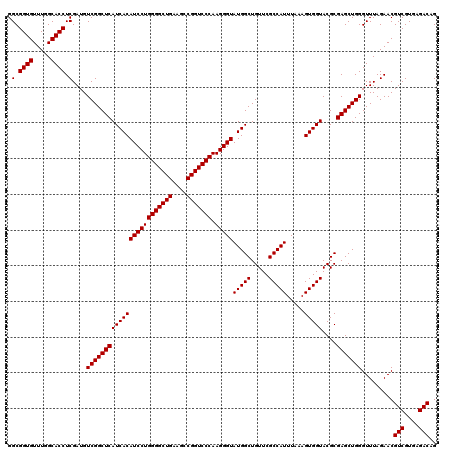
| Location | 4,789,176 – 4,789,296 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -48.70 |
| Consensus MFE | -48.70 |
| Energy contribution | -48.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789176 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >pfs_1 361 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >ppk_1 361 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >psp_1 358 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >pss_1 358 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >pst_1 358 120 - 1 GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... ( -48.70) >consensus GGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUU ((.((..((((((..((((((((...((.......))...))))))).)..)).((..((((......))))..((((((((((((....))))))).)))))..)))))))).)).... (-48.70 = -48.70 + -0.00)

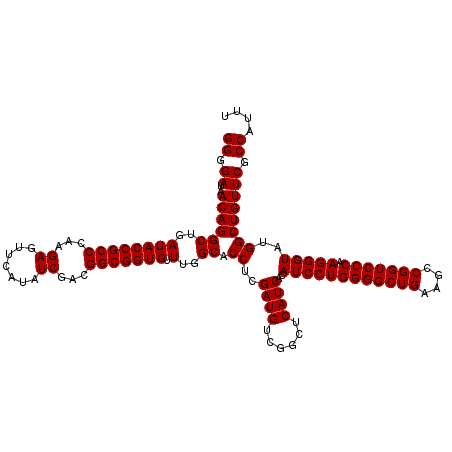
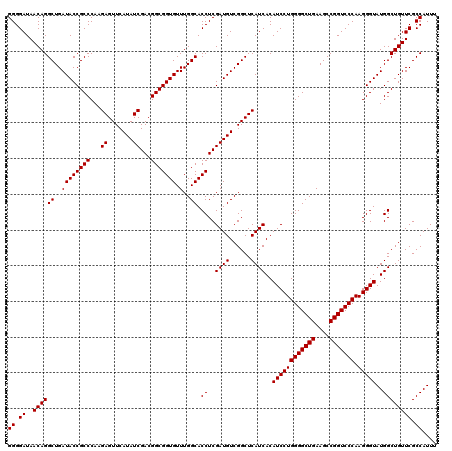
| Location | 4,789,256 – 4,789,376 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -37.70 |
| Energy contribution | -37.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789256 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >pfs_1 441 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >ppk_1 441 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >psp_1 438 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >pss_1 438 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >pst_1 438 120 - 1 CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... ( -37.70) >consensus CGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGAC .((((...(((....)))((((.....)))).(((((((((.......)))))))))))))....((((....(.((((..(((......((......)).)))..)))).).))))... (-37.70 = -37.70 + -0.00)

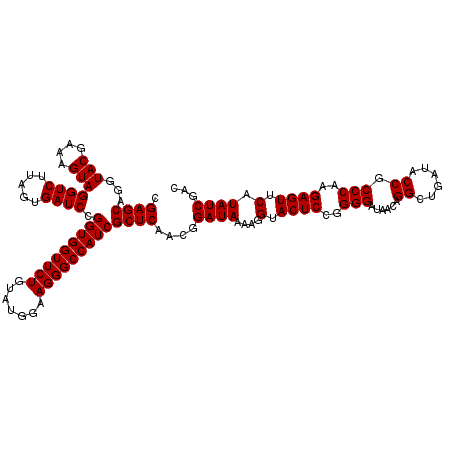
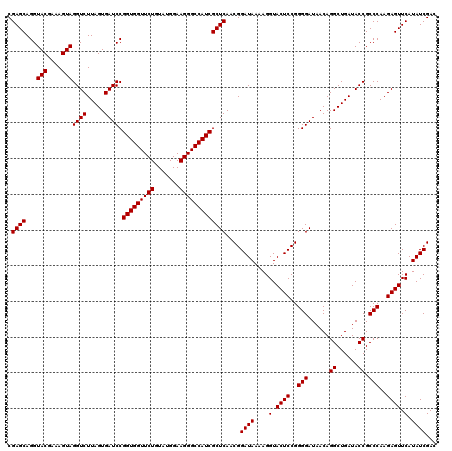
| Location | 4,789,296 – 4,789,416 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.56 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -35.50 |
| Energy contribution | -35.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789296 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >pel_1 481 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >ppk_1 481 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >psp_1 478 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >pss_1 478 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >pst_1 478 120 - 1 UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... ( -35.50) >consensus UAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCC .......((....))(..(((..((((..(((......)))..)))).(((....))).((((...(((..((((((((((.......)))))))))).)))..))))...)))..)... (-35.50 = -35.50 + 0.00)

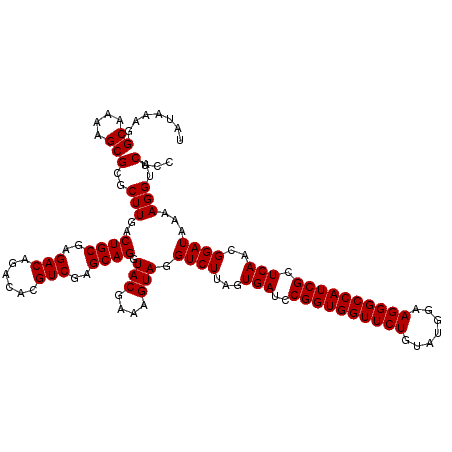
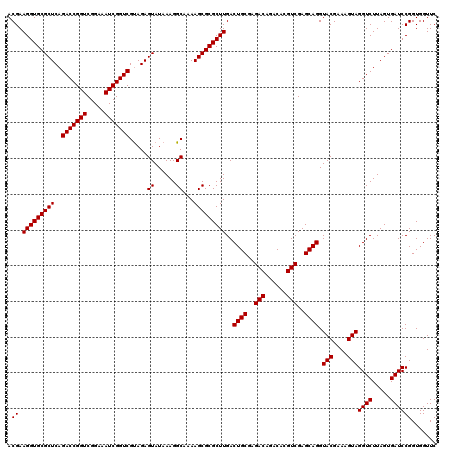
| Location | 4,789,336 – 4,789,456 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -40.30 |
| Energy contribution | -40.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789336 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.90) >pel_1 521 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.30) >ppk_1 521 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.30) >psp_1 518 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.30) >pss_1 518 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.30) >pst_1 518 120 - 1 ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... ( -40.30) >consensus ACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUC .((.(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))).(((....)))((((.....))))))....... (-40.30 = -40.30 + 0.00)

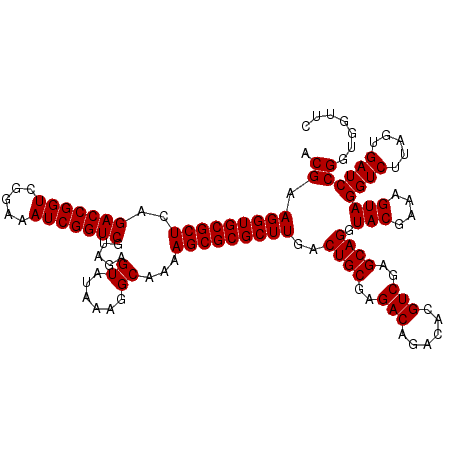
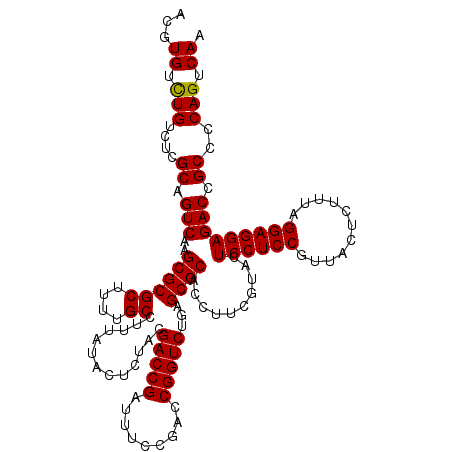
| Location | 4,789,376 – 4,789,496 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -34.02 |
| Energy contribution | -33.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.945799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789376 120 + 1 ACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAA ...((.(((....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))..))).)). ( -35.10) >pel_1 561 120 + 1 ACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAA .((((((((.(.....).)))))))).....((.......(((...(((((.........))))).))).........((.(((((((...........))))))))).))......... ( -32.10) >ppk_1 561 120 + 1 ACGUGUUUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAA .((((((((.(.....).)))))))).....((.......(((...(((((.........))))).))).........((.(((((((...........))))))))).))......... ( -32.10) >psp_1 558 120 + 1 ACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAA ...((.(((....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))..))).)). ( -35.10) >pss_1 558 120 + 1 ACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAA ...((.(((....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))..))).)). ( -35.10) >pst_1 558 120 + 1 ACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAA ...((.(((....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))..))).)). ( -35.10) >consensus ACGUGUCUGUCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAA ...((.(((....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).))..))).)). (-34.02 = -33.80 + -0.22)

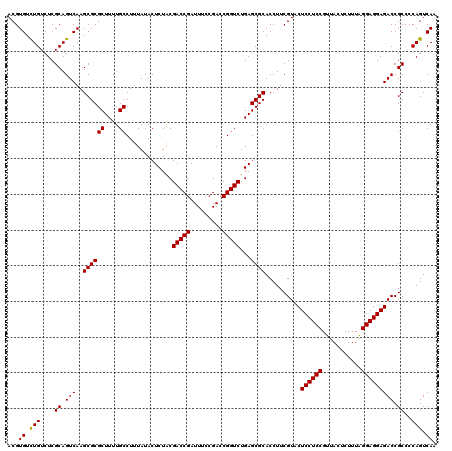
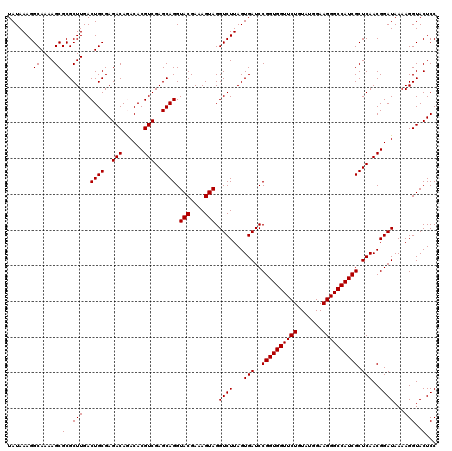
| Location | 4,789,376 – 4,789,496 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -42.30 |
| Energy contribution | -42.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789376 120 - 1 UUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGU ....(((..((((((((((((...........))))))......(((((((((..(((((((.....)))))))((...........))...)))))))))))))))....)))...... ( -44.30) >pel_1 561 120 - 1 UUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGU .........((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))............. ( -41.00) >ppk_1 561 120 - 1 UUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGU .........((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))............. ( -41.00) >psp_1 558 120 - 1 UUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGU ....(((..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))....)))...... ( -43.70) >pss_1 558 120 - 1 UUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGU ....(((..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))....)))...... ( -43.70) >pst_1 558 120 - 1 UUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGU ....(((..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))....)))...... ( -43.70) >consensus UUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGU .((.(((..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((......))...)))))))))))))))....))).))... (-42.30 = -42.63 + 0.33)

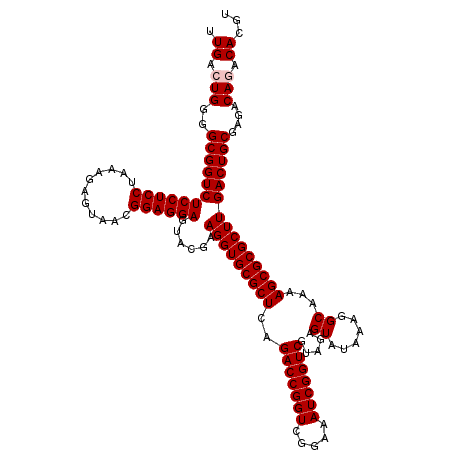
| Location | 4,789,416 – 4,789,536 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789416 120 + 1 CUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))...(((.....)))............))).))))..))))....)))).. ( -32.40) >pel_1 601 120 + 1 CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. ( -29.50) >ppk_1 601 120 + 1 CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. ( -29.50) >psp_1 598 120 + 1 CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. ( -30.10) >pss_1 598 120 + 1 CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. ( -30.10) >pst_1 598 120 + 1 CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. ( -30.10) >consensus CUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACC ..............((((.((((..((((.(((....((..(((((((...........)))))))..))....((.....)).............))).))))..))))....)))).. (-29.90 = -29.90 + 0.00)
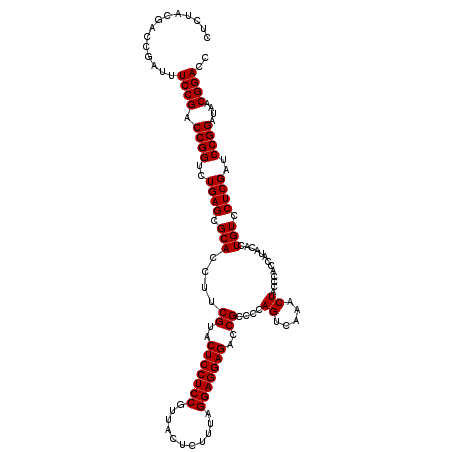
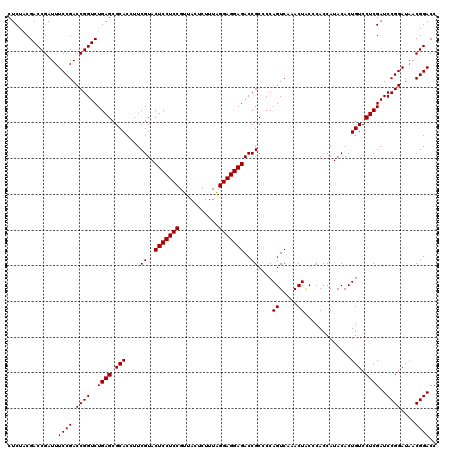
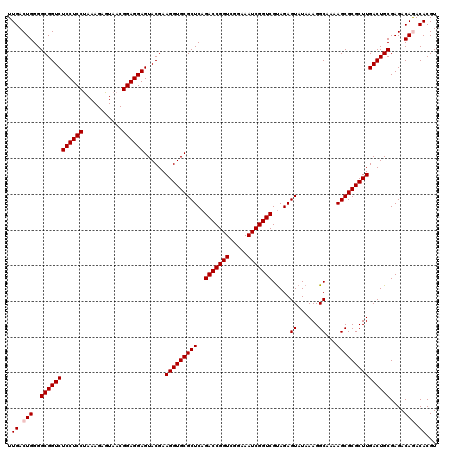
| Location | 4,789,416 – 4,789,536 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -42.47 |
| Consensus MFE | -40.98 |
| Energy contribution | -40.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789416 120 - 1 GGUCCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAG ((((((.....))))))........(((....(((((.....(((((...)))))((((((...........))))))((((....)))))))))(((((((.....))))))))))... ( -43.80) >pel_1 601 120 - 1 GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))........(.((((((.(((..(((((((((((((..(((((((...........)))))))(((....)))))))....))))))))).))).)))))).). ( -41.90) >ppk_1 601 120 - 1 GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))........(.((((((.(((..(((((((((((((..(((((((...........)))))))(((....)))))))....))))))))).))).)))))).). ( -41.90) >psp_1 598 120 - 1 GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))........(.((((((.(((..(((((((((((((..(((((((...........)))))))(((....)))))))....))))))))).))).)))))).). ( -42.40) >pss_1 598 120 - 1 GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))........(.((((((.(((..(((((((((((((..(((((((...........)))))))(((....)))))))....))))))))).))).)))))).). ( -42.40) >pst_1 598 120 - 1 GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))........(.((((((.(((..(((((((((((((..(((((((...........)))))))(((....)))))))....))))))))).))).)))))).). ( -42.40) >consensus GGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAG ((((((.....))))))((((((.((...((((.((....)).))))..)).)))((((((...........))))))((((....)))).))).(((((((.....)))))))...... (-40.98 = -40.98 + -0.00)

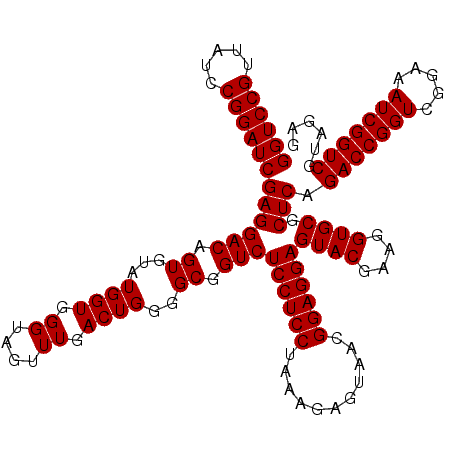
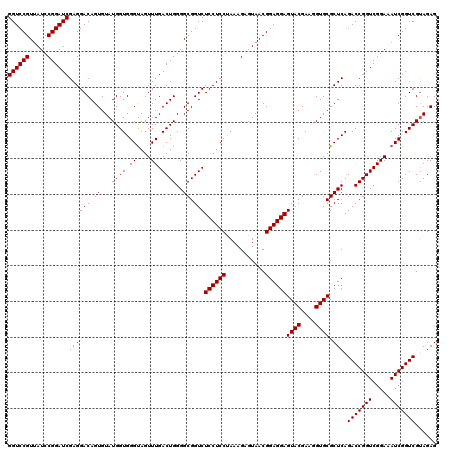
| Location | 4,789,456 – 4,789,576 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.72 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -33.74 |
| Energy contribution | -34.02 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789456 120 + 1 ACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCACCAUACACUGUCCUCGAUCCGGAUUACGGACCAGAGUUAGAACCUCAAGCAUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))((((((((((.....)))...........(((..(((..((((.....))))...))).)))................)))))))......... ( -36.70) >pel_1 641 120 + 1 ACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAGGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..................(((..((((.((((.....))))..)))).)))((((....))))))..)))))))......... ( -38.30) >ppk_1 641 120 + 1 ACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))......... ( -34.90) >psp_1 638 120 + 1 ACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))......... ( -35.50) >pss_1 638 120 + 1 ACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))......... ( -35.50) >pst_1 638 120 + 1 ACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))......... ( -35.50) >consensus ACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCACCAUACACUGUCCUCGAUCCGGAUAACGGACCUGAGUUAGAACCUCAAAGUUGCCAGGGUGGUAUUUCAAGG .(((((((...........)))))))(((((((..((..((((....................(.((((.....)))).)((((.......)))).))))))..)))))))......... (-33.74 = -34.02 + 0.28)

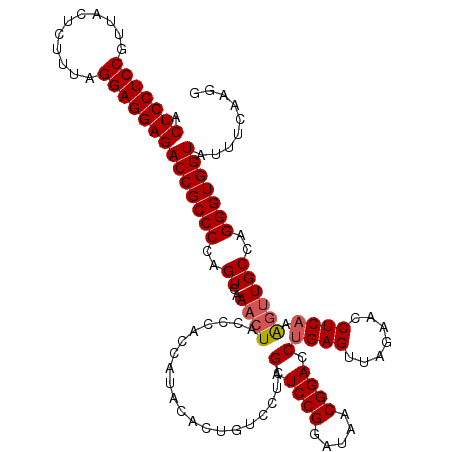
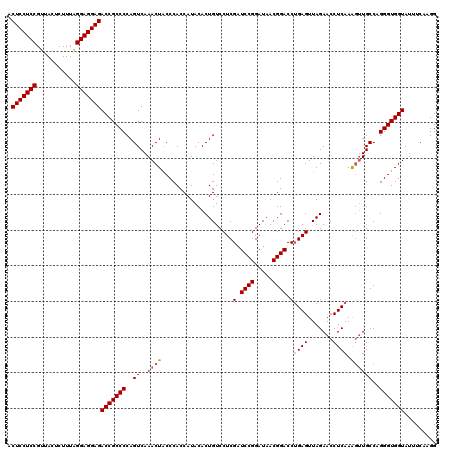
| Location | 4,789,496 – 4,789,616 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -45.16 |
| Energy contribution | -43.88 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.66 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
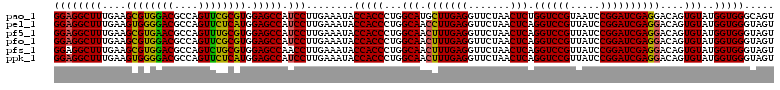
>pao_1 4789496 120 - 1 GGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCUGGUCCGUAAUCCGGAUCGAGGACAGUGUAUGGUGGGCAGU ((((((((....((((((((....)))))))).))))).)))........(((((...((((.((((((.......)))(((((((.....))))))))))...))))..)))))..... ( -46.80) >pel_1 681 120 - 1 GGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((((((((....((((((((....)))))))).))))).)))........(((((...(((.(((((((.......))).((((((.....))))))))))....)))..)))))..... ( -46.50) >pf5_1 678 120 - 1 GGAGGCUUUGAAGCGUGAACGCCAGUUUGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((((((((....(((..(((....)))..))).))))).)))........(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))..... ( -43.70) >pfo_1 681 120 - 1 GGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((((((((....((((((((....)))))))).))))).)))........(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))..... ( -45.60) >pfs_1 681 120 - 1 GGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((.(((((....(((..(((....)))..))).)))))..))........(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))..... ( -42.50) >ppk_1 681 120 - 1 GGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((((((((....((((((((....)))))))).))))).)))........(((((...(((.(((((((.......))))((((((.....))))))))).....)))..)))))..... ( -43.90) >consensus GGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCAGGUCCGUUAUCCGGAUCGAGGACAGUGUAUGGUGGGUAGU ((((((((....((((((((....)))))))).))))).)))........(((((...(((.(((((((.......))).((((((.....))))))))))....)))..)))))..... (-45.16 = -43.88 + -1.27)
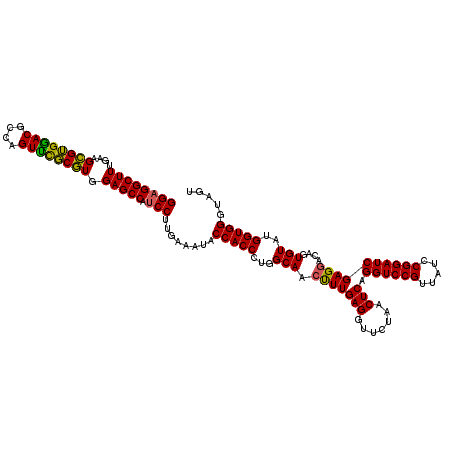
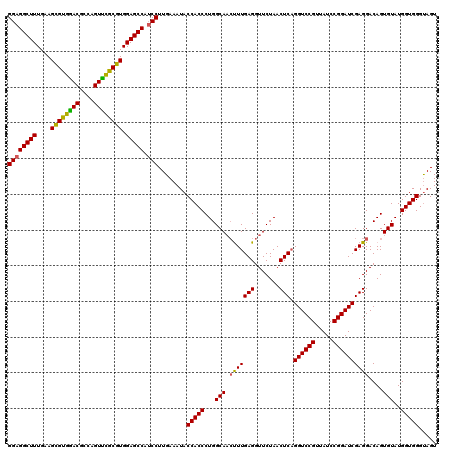
| Location | 4,789,536 – 4,789,656 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -41.38 |
| Consensus MFE | -41.74 |
| Energy contribution | -40.25 |
| Covariance contribution | -1.49 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.60 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 4789536 120 - 1 CUUUGCACUGGACUUUGAGCCUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAUGCUUGAGGUUCUAACUCU .........((((((..(((.((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).)))).)))..))))))....... ( -45.80) >pel_1 721 120 - 1 CUUUGCACUGGACUUUGAGCUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACCUUGAGGUUCUAACUCA .........((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))....... ( -41.60) >pf5_1 718 120 - 1 CUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGAACGCCAGUUUGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCA .........((((((..((.(((((.(.((.((.((...(((((((((....(((..(((....)))..))).))))).)))).....)))))).).))))).))..))))))....... ( -39.60) >pfo_1 721 120 - 1 CUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCA .........((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))....... ( -41.50) >ppk_1 721 120 - 1 CUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGUGGGGACGCCAGUUCUCAUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCA .........((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))....... ( -39.80) >pst_1 718 120 - 1 CUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUCUGCGUGGAGCCAACCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCA .........((((((..((.(((((.(((((....((((....(((((....(((..(((....)))..))).))))).)))).....)).)))...))))).))..))))))....... ( -40.00) >consensus CUUUGCACUGGACUUUGAAUUUGCUUGUGUAGGAUAGGUGGGAGGCUUUGAAGCGUGGACGCCAGUUCGCGUGGAGCCAUCCUUGAAAUACCACCCUGGCAACUUUGAGGUUCUAACUCA .........((((((..((.(((((.(.((.((.((...(((((((((....((((((((....)))))))).))))).)))).....)))))).).))))).))..))))))....... (-41.74 = -40.25 + -1.49)

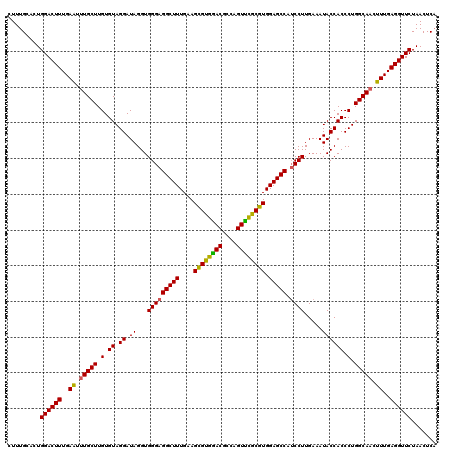
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:49:16 2007