| Sequence ID | pao_1 |
|---|---|
| Location | 4,785,689 – 4,785,808 |
| Length | 119 |
| Max. P | 0.841875 |

| Location | 4,785,689 – 4,785,808 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -37.28 |
| Energy contribution | -37.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
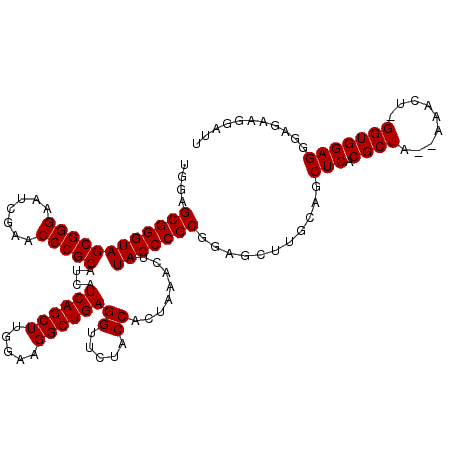
>pao_1 4785689 119 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUGAACUAUACCCGCGGAGCCCGAAGCUCACGCUUUUAAACU-GGUGGAGGGAGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))(((((.......))))).))))))).....((....(((.((((........-))))..)))....))... ( -43.40) >pel_1 1 117 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGGAGCUUGCGGCUCUCGCUG--AAACU-GGUGGAGGGAGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))(((((.....)))))((((.--.....-)))).............. ( -40.90) >pfo_1 1 118 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGGAGCUUGCAGCUCACGCUA--AAAUCUGGUGGAGGGAGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))((.....)).........))))))).((((.....)))).(((((--.....))))).............. ( -41.50) >pfs_1 1 116 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGGAGCUUGCAGCUCACGCUA--AAAU--GGUGGAGGGGGAAGGAUU ((..((((((..((.....)).))))))..))((((((.....))))))((((((.((.((..(((((....(((((((.....)))).)))..--..))--)))..))..)).)))))) ( -43.10) >pss_1 1 117 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCACAACUUGCAGCUCGCGCUA--AAUCU-GGUGGAGGGGGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))....(((.(..(((.(((((--....)-)))))))..).))).... ( -43.80) >pst_1 1 117 + 1 UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGCAAUUUGCAGCUCGCGCUA--AAUCU-GGUGGAGGGGGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))((.....))..(((.(((((--....)-)))))))........... ( -41.10) >consensus UGGAGCGGGUAGCGGGAAUCGAACCCGCAUCAUCAGCUUGGAAGGCUGAGGUUCUACCACUAAACUAUACCCGCGGAGCUUGCAGCUCACGCUA__AAACU_GGUGGAGGGAGAAGGAUU ....((((((((((((.......)))))....((((((.....))))))((.....)).........)))))))...........(((.((((.........)))))))........... (-37.28 = -37.28 + 0.00)


| Location | 4,785,689 – 4,785,808 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -33.94 |
| Energy contribution | -33.88 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
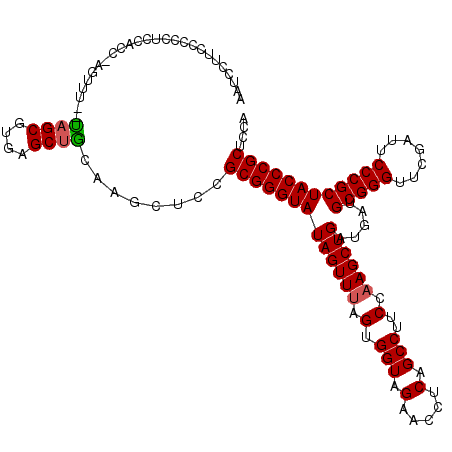
>pao_1 4785689 119 - 1 AAUCCUUCUCCCUCCACC-AGUUUAAAAGCGUGAGCUUCGGGCUCCGCGGGUAUAGUUCAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA ..................-(((((..((((....)))).)))))..(((((((..((((.......)))).(((((.......)))))....(((((.......)))))))))))).... ( -36.90) >pel_1 1 117 - 1 AAUCCUUCUCCCUCCACC-AGUUU--CAGCGAGAGCCGCAAGCUCCGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA ..................-(((((--(.((....)).).)))))..(((((((((((((.(.(((.(.....).)))..).)))))).....(((((.......)))))))))))).... ( -35.00) >pfo_1 1 118 - 1 AAUCCUUCUCCCUCCACCAGAUUU--UAGCGUGAGCUGCAAGCUCCGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA .....((((....((((.(((((.--..(((.((((.....)))))))......))))).)))).))))..(((((.......)))))((..((((((.((.....))..))))))..)) ( -36.80) >pfs_1 1 116 - 1 AAUCCUUCCCCCUCCACC--AUUU--UAGCGUGAGCUGCAAGCUCCGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA ..................--....--((((....))))........(((((((((((((.(.(((.(.....).)))..).)))))).....(((((.......)))))))))))).... ( -35.10) >pss_1 1 117 - 1 AAUCCUUCCCCCUCCACC-AGAUU--UAGCGCGAGCUGCAAGUUGUGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA .................(-((...--((((....))))....))).(((((((((((((.(.(((.(.....).)))..).)))))).....(((((.......)))))))))))).... ( -34.70) >pst_1 1 117 - 1 AAUCCUUCCCCCUCCACC-AGAUU--UAGCGCGAGCUGCAAAUUGCGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA ..................-.....--.(((....)))((.....))(((((((((((((.(.(((.(.....).)))..).)))))).....(((((.......)))))))))))).... ( -35.10) >consensus AAUCCUUCCCCCUCCACC_AGUUU__UAGCGUGAGCUGCAAGCUCCGCGGGUAUAGUUUAGUGGUAGAACCUCAGCCUUCCAAGCUGAUGAUGCGGGUUCGAUUCCCGCUACCCGCUCCA ..........................((((....))))........(((((((((((((.(.(((.(.....).)))..).)))))).....(((((.......)))))))))))).... (-33.94 = -33.88 + -0.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:48:55 2007