
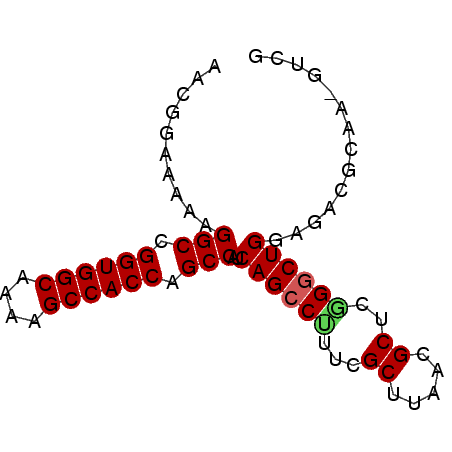
| Sequence ID | pao_1 |
|---|---|
| Location | 4,780,759 – 4,780,835 |
| Length | 76 |
| Max. P | 0.988671 |

| Location | 4,780,759 – 4,780,835 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | forward |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
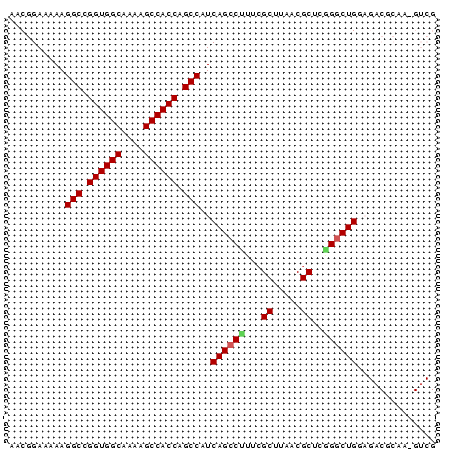
>pao_1 4780759 76 + 1 AACGGAAAAAGGCCGGUGGCAAAAGCCACCAGCCAUCAGCCUUUCGCUUGACGCUCGGGCUGUAGACGCAAGGUCG ..........(((.((((((....)))))).)))..((((((...((.....))..))))))..(((.....))). ( -30.80) >pel_1 1 76 + 1 AACGGAAAAAGGCCGGUGGCAUAAGCCACCAGCCAUCAGCCCUUCGCUUAACGCUCGGACUGGAGUCGCAAAGUCG ...((.....(((.((((((....)))))).))).....))....((((..(((((......))).))..)))).. ( -25.70) >pf5_1 1 76 + 1 AACGGAAAAAGGCCGGUGGCAAAAGCCACCAGCCAUCAGCCGUUCGCUUAACGCUUGGGCUGGAGACACAAAGUCG ..........(((.((((((....)))))).))).((((((....((.....))...)))))).(((.....))). ( -30.30) >pfo_1 1 62 + 1 AACGGAAAAAGGCCGGUGGCAAGAGCCACCAGCCAUCAGCCUGUCGCUUGACGCUCGGGCUG-------------- ..........(((.((((((....)))))).)))..(((((((..((.....)).)))))))-------------- ( -30.40) >pfs_1 1 76 + 1 AACGGAAAAAGGCCGGUGGCAAGAGCCACCAGCCAUCAGCCUUCAGCUUAACGCUUGGGCUGGAGACGCAAGGUCG ..........(((.((((((....)))))).))).(((((((..(((.....))).))))))).(((.....))). ( -34.10) >ppk_1 1 62 + 1 AACGGAAAAAGGCCGGUGGCAUAAGCCACCAGCCAUCAGCCUUUCGCUCAACGCUCAGGCUG-------------- ..........(((.((((((....)))))).)))..((((((...((.....))..))))))-------------- ( -27.70) >consensus AACGGAAAAAGGCCGGUGGCAAAAGCCACCAGCCAUCAGCCUUUCGCUUAACGCUCGGGCUGGAGACGCAA_GUCG ..........(((.((((((....)))))).)))..((((((...((.....))..)))))).............. (-27.39 = -27.50 + 0.11)

| Location | 4,780,759 – 4,780,835 |
|---|---|
| Length | 76 |
| Sequences | 6 |
| Columns | 76 |
| Reading direction | reverse |
| Mean pairwise identity | 82.59 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -32.24 |
| Energy contribution | -32.10 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.35 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
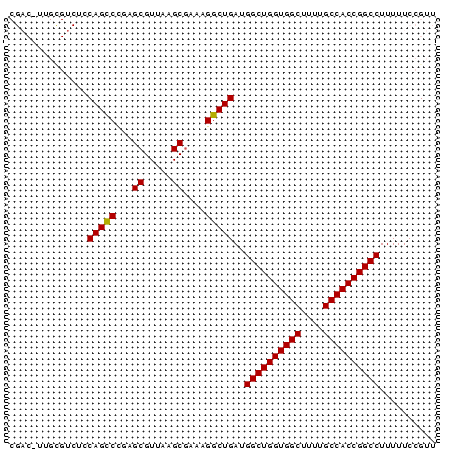
>pao_1 4780759 76 - 1 CGACCUUGCGUCUACAGCCCGAGCGUCAAGCGAAAGGCUGAUGGCUGGUGGCUUUUGCCACCGGCCUUUUUCCGUU .(((.....)))..(((((...((.....))....)))))..((((((((((....)))))))))).......... ( -34.40) >pel_1 1 76 - 1 CGACUUUGCGACUCCAGUCCGAGCGUUAAGCGAAGGGCUGAUGGCUGGUGGCUUAUGCCACCGGCCUUUUUCCGUU .......(((....((((((..((.....))...))))))..((((((((((....))))))))))......))). ( -36.40) >pf5_1 1 76 - 1 CGACUUUGUGUCUCCAGCCCAAGCGUUAAGCGAACGGCUGAUGGCUGGUGGCUUUUGCCACCGGCCUUUUUCCGUU .(((.....)))..(((((...((.....))....)))))..((((((((((....)))))))))).......... ( -35.30) >pfo_1 1 62 - 1 --------------CAGCCCGAGCGUCAAGCGACAGGCUGAUGGCUGGUGGCUCUUGCCACCGGCCUUUUUCCGUU --------------(((((.(.((.....))..).)))))..((((((((((....)))))))))).......... ( -32.50) >pfs_1 1 76 - 1 CGACCUUGCGUCUCCAGCCCAAGCGUUAAGCUGAAGGCUGAUGGCUGGUGGCUCUUGCCACCGGCCUUUUUCCGUU .(((.....)))..(((((..(((.....)))...)))))..((((((((((....)))))))))).......... ( -36.00) >ppk_1 1 62 - 1 --------------CAGCCUGAGCGUUGAGCGAAAGGCUGAUGGCUGGUGGCUUAUGCCACCGGCCUUUUUCCGUU --------------((((((..((.....))...))))))..((((((((((....)))))))))).......... ( -35.40) >consensus CGAC_UUGCGUCUCCAGCCCGAGCGUUAAGCGAAAGGCUGAUGGCUGGUGGCUUUUGCCACCGGCCUUUUUCCGUU ..............(((((...((.....))....)))))..((((((((((....)))))))))).......... (-32.24 = -32.10 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:48:37 2007