
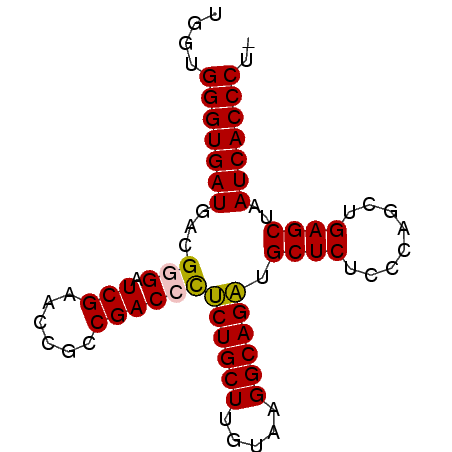
| Sequence ID | pao_1 |
|---|---|
| Location | 3,650,816 – 3,650,894 |
| Length | 78 |
| Max. P | 0.999848 |

| Location | 3,650,816 – 3,650,894 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.73 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
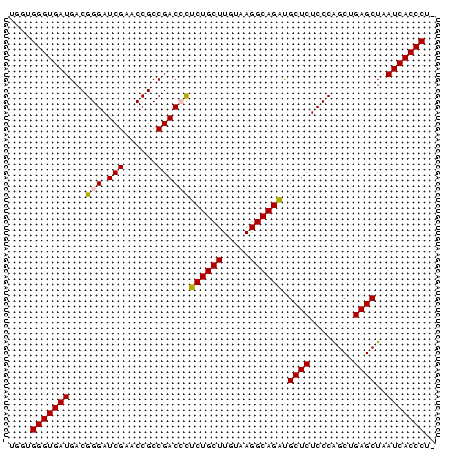
>pao_1 3650816 78 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCACCCUU ....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).. ( -31.30) >pel_1 1 76 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCGGCUGAGCUAAUCACCC-- ....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))-- ( -30.10) >pf5_1 1 78 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACAUUCUGCUUGUAAGGCAGACGCUCUCCCGGCUGAGCUAAUCACCCUU ....((((((((.(((.......))).).....((((((.....)))))).((((........))))..))))))).. ( -27.70) >pfo_1 1 76 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACAUUCUGCUUGUAAGGCAGACGCUCUCCCAGCUGAGCUAAUCACCC-- ....((((((((.(((.......))).).....((((((.....)))))).((((........))))..)))))))-- ( -26.50) >ppk_1 1 77 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCGGCUGAGCUAAUCACCCU- ....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).- ( -31.30) >pst_1 1 78 + 1 UGGUGGGUGAUGACGGGAUCGAACCGCCGACCCGCUGCUUGUAAGGCAGCUGCUCUCCCAGCUGAGCUAAUCACCCUU ....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).. ( -34.10) >consensus UGGUGGGUGAUGACGGGAUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCACCCU_ ....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).. (-28.90 = -28.73 + -0.16)

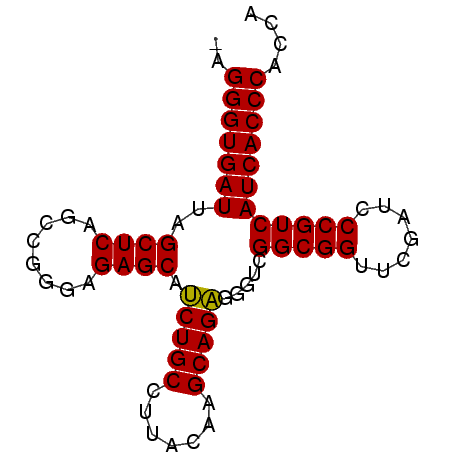
| Location | 3,650,816 – 3,650,894 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 94.85 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -29.96 |
| Energy contribution | -29.68 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
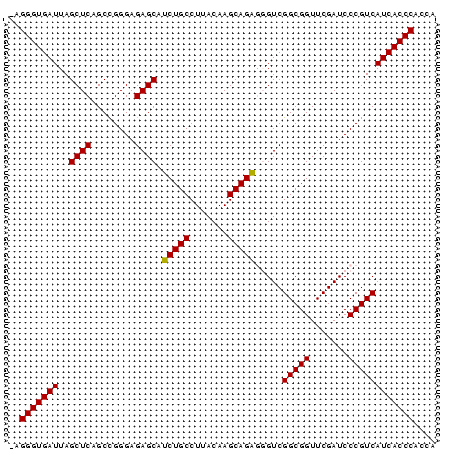
>pao_1 3650816 78 - 1 AAGGGUGAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAUCCCGUCAUCACCCACCA ..(((((((..((((........)))).(((((.......)))))(((((((....)))))))....))))))).... ( -30.70) >pel_1 1 76 - 1 --GGGUGAUUAGCUCAGCCGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAUCCCGUCAUCACCCACCA --(((((((..((((........)))).(((((.......)))))(((((((....)))))))....))))))).... ( -30.50) >pf5_1 1 78 - 1 AAGGGUGAUUAGCUCAGCCGGGAGAGCGUCUGCCUUACAAGCAGAAUGUCGGCGGUUCGAUCCCGUCAUCACCCACCA ..(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).... ( -29.30) >pfo_1 1 76 - 1 --GGGUGAUUAGCUCAGCUGGGAGAGCGUCUGCCUUACAAGCAGAAUGUCGGCGGUUCGAUCCCGUCAUCACCCACCA --(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).... ( -29.10) >ppk_1 1 77 - 1 -AGGGUGAUUAGCUCAGCCGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAUCCCGUCAUCACCCACCA -.(((((((..((((........)))).(((((.......)))))(((((((....)))))))....))))))).... ( -30.70) >pst_1 1 78 - 1 AAGGGUGAUUAGCUCAGCUGGGAGAGCAGCUGCCUUACAAGCAGCGGGUCGGCGGUUCGAUCCCGUCAUCACCCACCA ..(((((((..((((........)))).(((((.......)))))(((((((....)))))))....))))))).... ( -33.10) >consensus _AGGGUGAUUAGCUCAGCCGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAUCCCGUCAUCACCCACCA ..(((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).... (-29.96 = -29.68 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:48:26 2007