
| Sequence ID | pao_1 |
|---|---|
| Location | 3,395,191 – 3,395,313 |
| Length | 122 |
| Max. P | 0.917750 |

| Location | 3,395,191 – 3,395,308 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -48.57 |
| Consensus MFE | -45.64 |
| Energy contribution | -45.87 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3395191 117 + 1 CGGAGCGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG---AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGC (((((((..((((.((...)).)))).(((((((...))))))).....(((((.......))))))))))))..........(---(((((((((((.....))))))))))))..... ( -51.30) >pau_1 1 117 + 1 CGGAGCGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG---AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGC (((((((..((((.((...)).)))).(((((((...))))))).....(((((.......))))))))))))..........(---(((((((((((.....))))))))))))..... ( -51.30) >pel_1 1 117 + 1 CGGAGUGUAGCGCAGCCAGGUAGCGCGUCUCGUUCGGGACGAGAAGGCCGCAGGUUCGAAUCCUGUCUCUCCGACCAAAUCCCGUAAAAAAACCCGCCUCGA--GCGGGUUUUUU-UUGC (((((.((((((.((((.(((..(.((((((....)))))).)...)))...))))))....))).).)))))..........((((((((((((((.....--))))).)))))-)))) ( -43.10) >consensus CGGAGCGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG___AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGC (((((((..((((.((...)).)))).(((((((...))))))).....(((((.......)))))))))))).........((...(((((((((((.....)))))))))))...)). (-45.64 = -45.87 + 0.22)

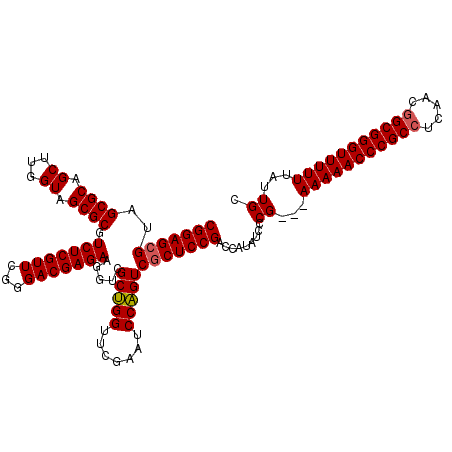
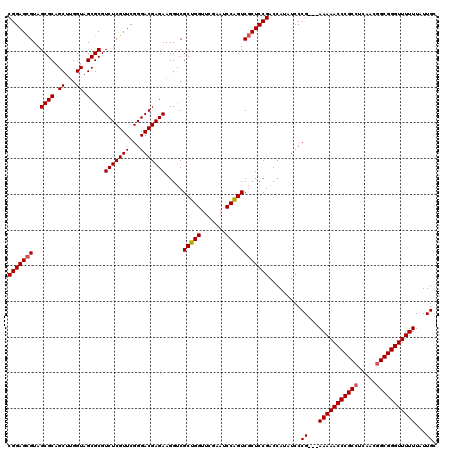
| Location | 3,395,191 – 3,395,308 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -45.63 |
| Consensus MFE | -43.33 |
| Energy contribution | -43.00 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3395191 117 - 1 GCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU---CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACGCUCCG ......(((((((((((.....)))))))))))---...........(((((((.((((.......))))......((((((.....)))))).((((.........))))..))))))) ( -46.90) >pau_1 1 117 - 1 GCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU---CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACGCUCCG ......(((((((((((.....)))))))))))---...........(((((((.((((.......))))......((((((.....)))))).((((.........))))..))))))) ( -46.90) >pel_1 1 117 - 1 GCAA-AAAAAACCCGC--UCGAGGCGGGUUUUUUUACGGGAUUUGGUCGGAGAGACAGGAUUCGAACCUGCGGCCUUCUCGUCCCGAACGAGACGCGCUACCUGGCUGCGCUACACUCCG ...(-(((((((((((--.....)))))))))))).(((..(.(((.((.((...((((...((......))(((.((((((.....)))))).).))..)))).)).))))).)..))) ( -43.10) >consensus GCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU___CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACGCUCCG ......(((((((((((.....)))))))))))...((((((..(((((......((((.......))))))))).....))))))...(((..((((.........))))....))).. (-43.33 = -43.00 + -0.33)

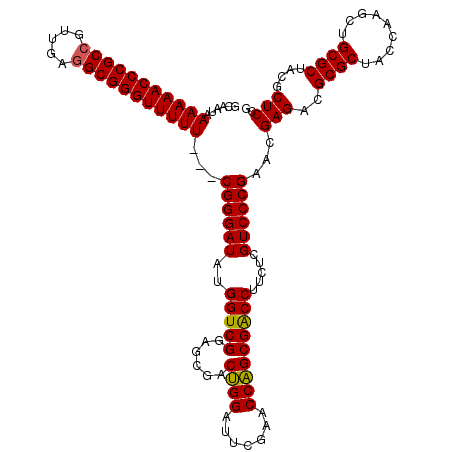
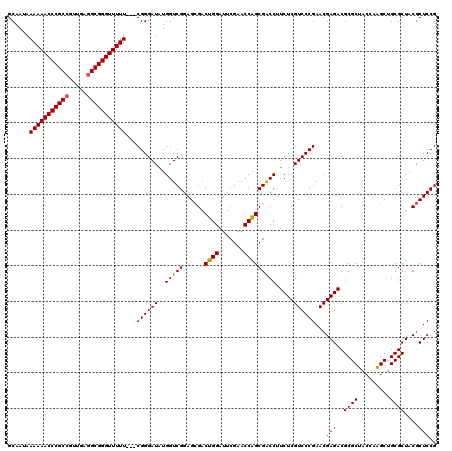
| Location | 3,395,196 – 3,395,313 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -48.17 |
| Consensus MFE | -43.70 |
| Energy contribution | -43.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3395196 117 + 1 CGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG---AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGCCUGCG (((((.((((.((((.((((((((((....))))))........(((((.......))))))))))))).).......(---(((((((((((.....))))))))))))..)))))))) ( -51.00) >pau_1 6 117 + 1 CGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG---AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGCCUGCG (((((.((((.((((.((((((((((....))))))........(((((.......))))))))))))).).......(---(((((((((((.....))))))))))))..)))))))) ( -51.00) >pel_1 6 117 + 1 UGUAGCGCAGCCAGGUAGCGCGUCUCGUUCGGGACGAGAAGGCCGCAGGUUCGAAUCCUGUCUCUCCGACCAAAUCCCGUAAAAAAACCCGCCUCGA--GCGGGUUUUUU-UUGCCUGCG .((((.(((....(((..(.((((((....)))))).)...)))(((((.......)))))...................((((((((((((.....--)))))))))))-)))))))). ( -42.50) >consensus CGUAGCGCAGCUUGGUAGCGCGUCUCGUUCGGGACGAGAAGGUCGCUGGUUCGAAUCCAGUCGCUCCGACCAUAUCCCG___AAAAACCCGCCUCAACGGCGGGUUUUUUAUUGCCUGCG (((((.((((...(((..(.((((((....)))))).)..((..(((((.......)))))....)).)))...........(((((((((((.....)))))))))))..))))))))) (-43.70 = -43.37 + -0.33)

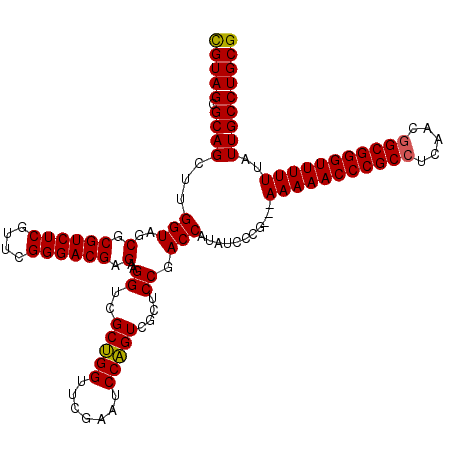
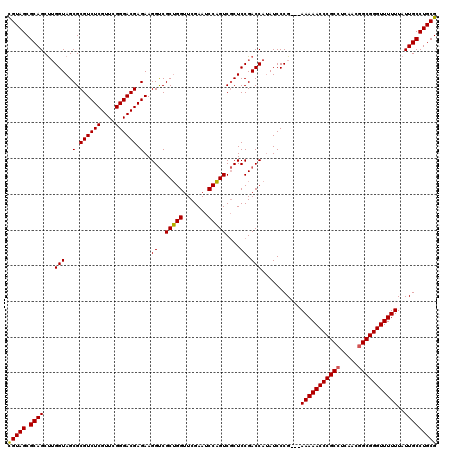
| Location | 3,395,196 – 3,395,313 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -43.92 |
| Energy contribution | -43.37 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3395196 117 - 1 CGCAGGCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU---CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACG ((((.((.....((((((((((.....))))))))))(---((((((..(((((......((((.......))))))))).....))))))).......))(((....)))))))..... ( -46.20) >pau_1 6 117 - 1 CGCAGGCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU---CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACG ((((.((.....((((((((((.....))))))))))(---((((((..(((((......((((.......))))))))).....))))))).......))(((....)))))))..... ( -46.20) >pel_1 6 117 - 1 CGCAGGCAA-AAAAAACCCGC--UCGAGGCGGGUUUUUUUACGGGAUUUGGUCGGAGAGACAGGAUUCGAACCUGCGGCCUUCUCGUCCCGAACGAGACGCGCUACCUGGCUGCGCUACA ((((((..(-(((((((((((--.....))))))))))))...(((((..(((.....)))..)))))...))))))....((((((.....)))))).((((.........)))).... ( -48.30) >consensus CGCAGGCAAUAAAAAACCCGCCGUUGAGGCGGGUUUUU___CGGGAUAUGGUCGGAGCGACUGGAUUCGAACCAGCGACCUUCUCGUCCCGAACGAGACGCGCUACCAAGCUGCGCUACG ((((.((....(((((((((((.....)))))))))))...((((((..(((((......((((.......))))))))).....))))))........))(((....)))))))..... (-43.92 = -43.37 + -0.55)

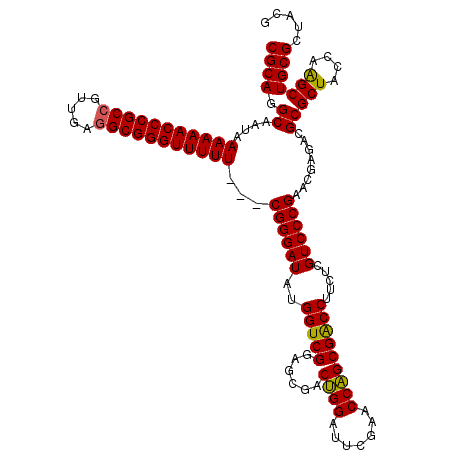
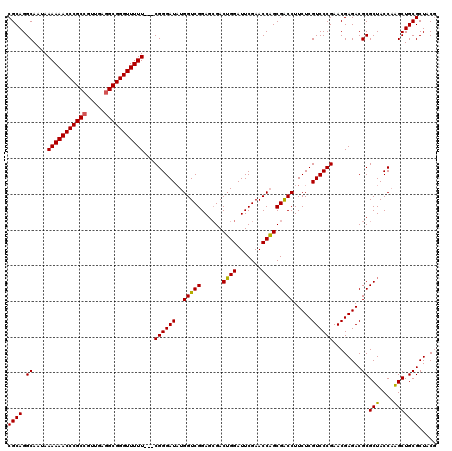
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:48:05 2007