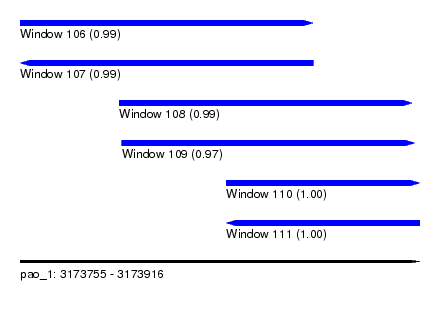
| Sequence ID | pao_1 |
|---|---|
| Location | 3,173,755 – 3,173,916 |
| Length | 161 |
| Max. P | 0.999959 |

| Location | 3,173,755 – 3,173,873 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -42.10 |
| Energy contribution | -42.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173755 118 + 1 GAAUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA--UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG ......((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))..--(((.(((((....))))))))...........)). ( -42.20) >pst_1 1 120 + 1 GAAUAUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG .((((.((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))....(((.(((((....))))))))...)).)))).... ( -43.10) >pel_1 1 114 + 1 -----UGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA-AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG -----.((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))..-.(((.(((((....))))))))...........)). ( -42.20) >pf5_1 1 119 + 1 GAAUUUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAA-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG ......((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))...-(((.(((((....))))))))...........)). ( -42.20) >pfs_1 1 119 + 1 GAAUAUGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA-AUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG .((((.((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))..-.(((.(((((....))))))))...)).)))).... ( -43.10) >ppk_1 1 114 + 1 -----UGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUU-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG -----.((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..))))))).((-(((.(((((....)))))))))).........)). ( -44.10) >consensus GAAU_UGGAGCGGGAAACGAGACUCGAACUCGCGACCCCGACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA__UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCG ......((.(((((((..(((.......)))(((((((((....)))...))))))((((........))))..)))))))......(.(((((....))))).)............)). (-42.10 = -42.10 + 0.00)

| Location | 3,173,755 – 3,173,873 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -47.50 |
| Energy contribution | -47.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173755 118 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA--UUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAUUC ........(((((....(((((....)))))))).--..(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).))...... ( -47.50) >pst_1 1 120 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAUAUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAUAUUC ........(((((....(((((....)))))))).....(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).))...... ( -47.50) >pel_1 1 114 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAU-UUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCA----- ........(((((....(((((....))))))))..-..(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).)).----- ( -47.50) >pf5_1 1 119 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA-UUUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAAAUUC ........(((((....(((((....)))))))).-...(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).))...... ( -47.50) >pfs_1 1 119 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCAU-UUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCAUAUUC ........(((((....(((((....))))))))..-..(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).))...... ( -47.50) >ppk_1 1 114 - 1 CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA-AAUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCA----- ........(((((....(((((....)))))))).-...(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).)).----- ( -47.50) >consensus CGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA__UUGCGGGAAUAGCUCAGUUGGUAGAGCACGACCUUGCCAAGGUCGGGGUCGCGAGUUCGAGUCUCGUUUCCCGCUCCA_AUUC ........(((((....(((((....)))))))).....(((((((..((((........))))..((((((((....).)))))))(((((.......)))))))))))).))...... (-47.50 = -47.50 + 0.00)


| Location | 3,173,795 – 3,173,913 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -45.50 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.987995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173795 118 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA--UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).))..--.)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). ( -46.80) >pst_1 41 120 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).)).....)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). ( -45.50) >pf5_1 41 119 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAA-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).))...-.)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). ( -46.10) >ppk_1 36 119 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUU-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA ......((..((....((((........))))....)).)).((-(((.(((((....))))))))))(((((...((((((.......))))))(((.(((....))))))..))))). ( -46.70) >psp_1 41 120 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).)).....)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). ( -45.50) >pss_1 41 120 + 1 ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).)).....)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). ( -45.50) >consensus ACCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAU_UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGA .((...((..((....((((........))))....)).)).....)).(((((....))))).....(((((...((((((.......))))))(((.(((....))))))..))))). (-45.50 = -45.50 + 0.00)

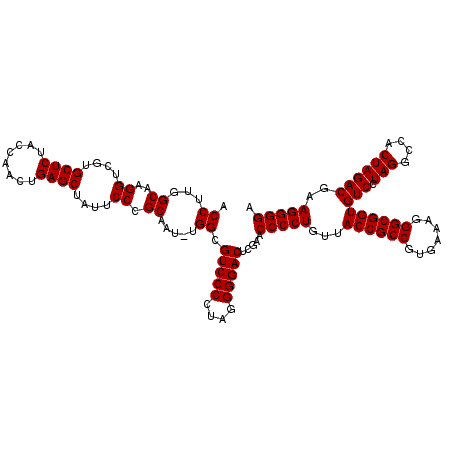
| Location | 3,173,796 – 3,173,914 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -45.50 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.967020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173796 118 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAA--UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).))..--.)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) ( -46.80) >pst_1 42 120 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).)).....)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) ( -45.50) >pf5_1 42 119 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAA-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).))...-.)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) ( -46.10) >ppk_1 37 119 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAUU-UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC .....((..((....((((........))))....)).)).((-(((.(((((....))))))))))(((((...((((((.......))))))(((.(((....))))))..))))).. ( -46.70) >psp_1 42 120 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).)).....)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) ( -45.50) >pss_1 42 120 + 1 CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAUAUGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).)).....)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) ( -45.50) >consensus CCUUGGCAAGGUCGUGCUCUACCAACUGAGCUAUUCCCGCAAU_UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC ((...((..((....((((........))))....)).)).....)).((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))) (-45.50 = -45.50 + 0.00)
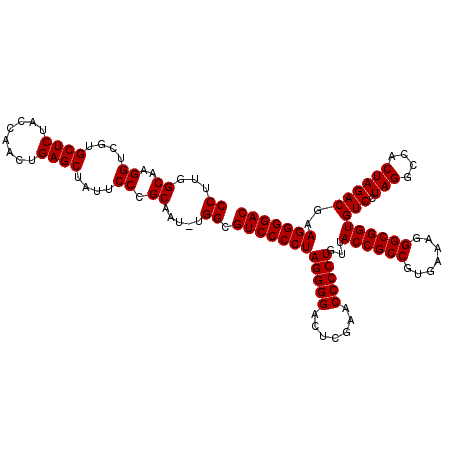

| Location | 3,173,838 – 3,173,916 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 98.88 |
| Mean single sequence MFE | -38.48 |
| Consensus MFE | -37.25 |
| Energy contribution | -37.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.89 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173838 78 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGC ..((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))) ( -41.00) >pau_1 1 77 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG- ...(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))- ( -37.80) >pel_1 1 77 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG- ...(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))- ( -37.80) >pfs_1 1 76 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGAC-- ....((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))-- ( -35.50) >ppk_1 1 77 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG- ...(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))- ( -37.80) >pss_1 1 78 + 1 UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACGC ..((((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..))))))))) ( -41.00) >consensus UGGCGUCCCCUAGGGGACUCGAACCCCUGUUACCGCCGUGAAAGGGCGGUGUCCUAGGCCACUAGACGAAGGGGACG_ ...(((((((((((((.......)))))...((((((.......))))))(((.(((....))))))..)))))))). (-37.25 = -37.42 + 0.17)

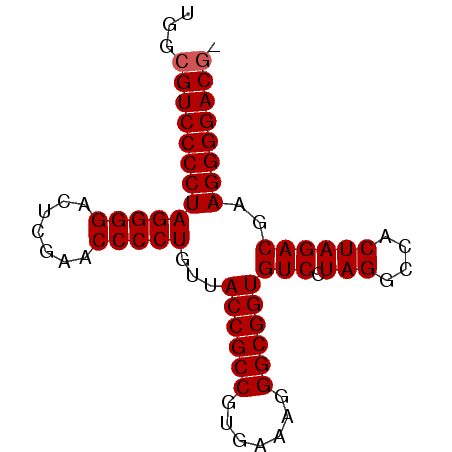
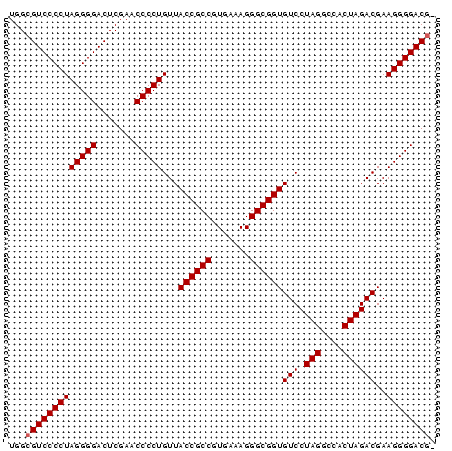
| Location | 3,173,838 – 3,173,916 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 98.88 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -35.45 |
| Energy contribution | -35.62 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 3173838 78 - 1 GCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA (((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))).. ( -39.90) >pau_1 1 77 - 1 -CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA -((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))... ( -36.10) >pel_1 1 77 - 1 -CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA -((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))... ( -36.10) >pfs_1 1 76 - 1 --GUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA --(((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))).... ( -35.00) >ppk_1 1 77 - 1 -CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA -((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))... ( -36.10) >pss_1 1 78 - 1 GCGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA (((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))).. ( -39.90) >consensus _CGUCCCCUUCGUCUAGUGGCCUAGGACACCGCCCUUUCACGGCGGUAACAGGGGUUCGAGUCCCCUAGGGGACGCCA .((((((((..((((((....))).)))((((((.......))))))...(((((.......)))))))))))))... (-35.45 = -35.62 + 0.17)

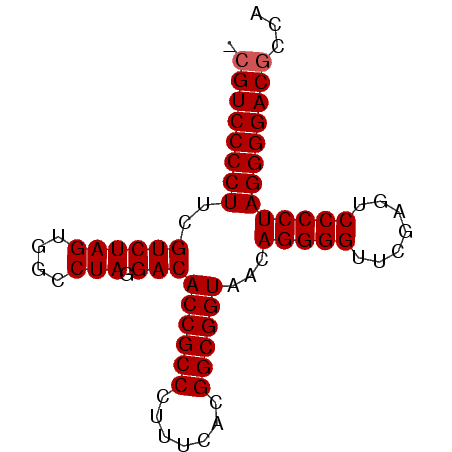
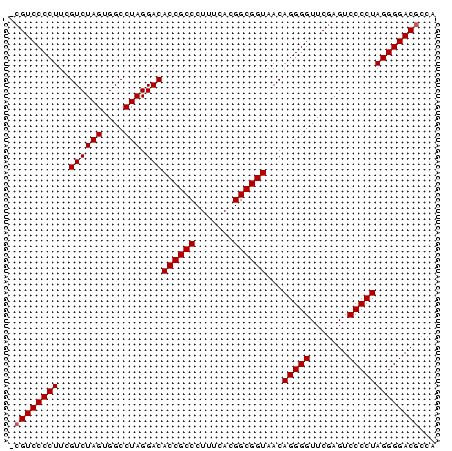
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:47:54 2007