
| Sequence ID | pao_1 |
|---|---|
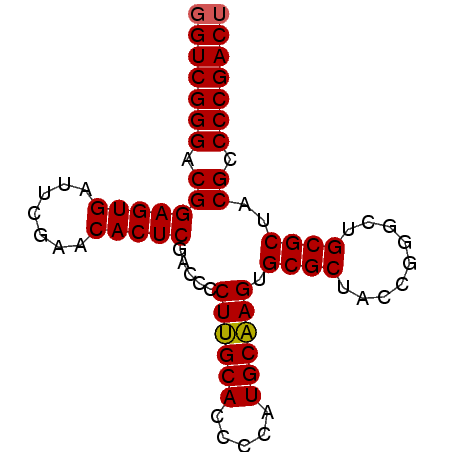
| Location | 3,099,308 – 3,099,387 |
| Length | 79 |
| Max. P | 0.999791 |

| Location | 3,099,308 – 3,099,387 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 96.88 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.48 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
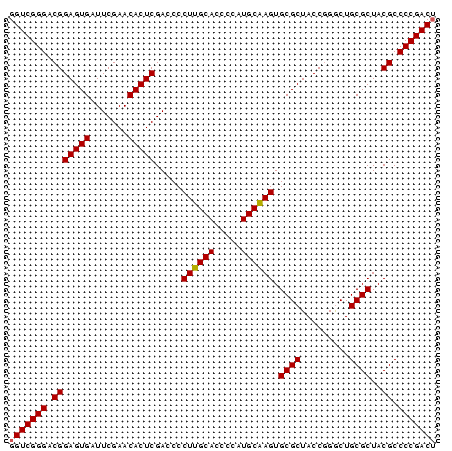
>pao_1 3099308 79 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUUGCACCCCAUGCAAGUGCGCUACCAGGCUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) ( -31.70) >pst_1 1 79 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUAGCACCCCAUGCUAGUGCGCUACCGGACUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) ( -32.10) >pau_1 1 79 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUUGCACCCCAUGCAAGUGCGCUACCAGGCUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) ( -31.70) >pel_1 1 78 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUUGCACCCCAUGCAAGUGCGCUACCGGGCUGCGCUACGCCCCGAC- .((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))- ( -30.10) >ppk_1 1 79 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUUGCACCCCAUGCAAGUGCGCUACCGGGCUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) ( -31.70) >pss_1 1 79 + 1 GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUAGCACCCCAUGCUAGUGCGCUACCGGACUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) ( -32.10) >consensus GGUCGGGACGGAGUGAUUCGAACACUCGACCCCUUGCACCCCAUGCAAGUGCGCUACCGGGCUGCGCUACGCCCCGACU (((((((.(((((((.......))))).....((((((.....)))))).((((.........))))..)).))))))) (-31.76 = -31.48 + -0.28)

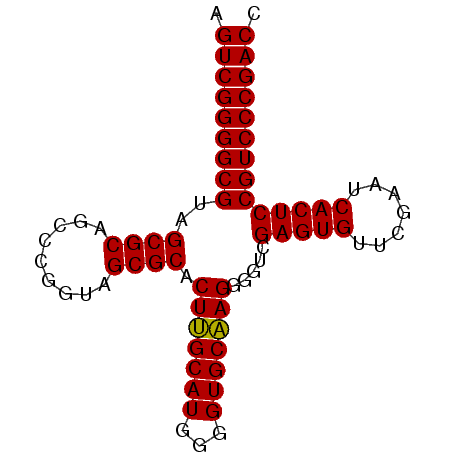
| Location | 3,099,308 – 3,099,387 |
|---|---|
| Length | 79 |
| Sequences | 6 |
| Columns | 79 |
| Reading direction | reverse |
| Mean pairwise identity | 96.88 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -36.74 |
| Energy contribution | -36.30 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
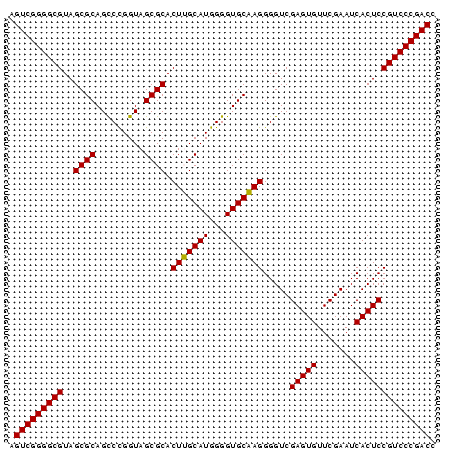
>pao_1 3099308 79 - 1 AGUCGGGGCGUAGCGCAGCCUGGUAGCGCACUUGCAUGGGGUGCAAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.((...)).)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.80) >pst_1 1 79 - 1 AGUCGGGGCGUAGCGCAGUCCGGUAGCGCACUAGCAUGGGGUGCUAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.........)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.60) >pau_1 1 79 - 1 AGUCGGGGCGUAGCGCAGCCUGGUAGCGCACUUGCAUGGGGUGCAAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.((...)).)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.80) >pel_1 1 78 - 1 -GUCGGGGCGUAGCGCAGCCCGGUAGCGCACUUGCAUGGGGUGCAAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC -(((((((((..((((.((...)).)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.60) >ppk_1 1 79 - 1 AGUCGGGGCGUAGCGCAGCCCGGUAGCGCACUUGCAUGGGGUGCAAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.((...)).)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.80) >pss_1 1 79 - 1 AGUCGGGGCGUAGCGCAGUCCGGUAGCGCACUAGCAUGGGGUGCUAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.........)))).(((((((...))))))).....(((((.......)))))))))))))). ( -36.60) >consensus AGUCGGGGCGUAGCGCAGCCCGGUAGCGCACUUGCAUGGGGUGCAAGGGGUCGAGUGUUCGAAUCACUCCGUCCCGACC .(((((((((..((((.........)))).(((((((...))))))).....(((((.......)))))))))))))). (-36.74 = -36.30 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:47:42 2007