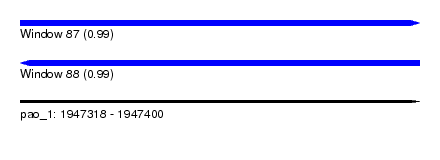
| Sequence ID | pao_1 |
|---|---|
| Location | 1,947,318 – 1,947,400 |
| Length | 82 |
| Max. P | 0.993874 |

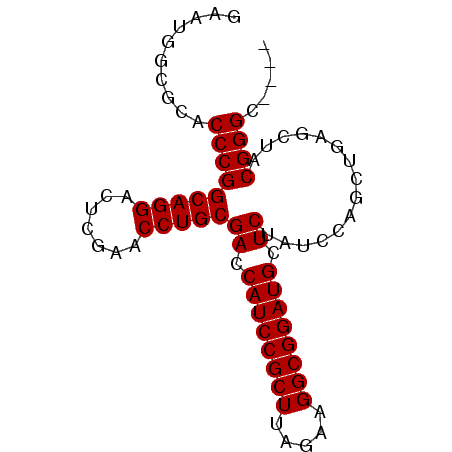
| Location | 1,947,318 – 1,947,400 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
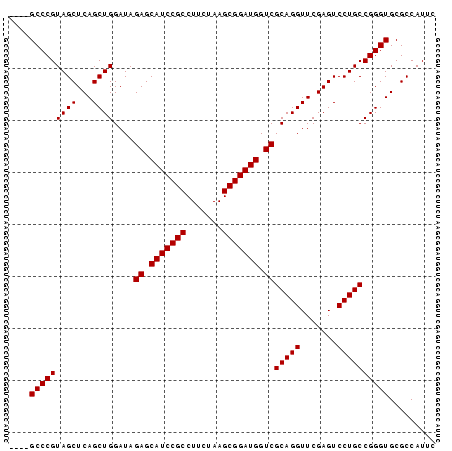
>pao_1 1947318 82 + 1 AAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUC ..(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))....... ( -38.10) >pst_1 1 77 + 1 ----GCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUU- ----(((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))........- ( -31.60) >pau_1 1 82 + 1 AAGCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUC ..(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))....... ( -38.10) >pfo_1 1 79 + 1 --GCGCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUU- --(((((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))))......- ( -37.90) >psp_1 1 78 + 1 ----GCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUC ----(((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))......... ( -31.60) >pss_1 1 78 + 1 ----GCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUC ----(((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))......... ( -31.60) >consensus ____GCCCGUAGCUCAGCUGGAUAGAGCAUCCGCCUUCUAAGCGGAUGGUCGCAGGUUCGAGUCCUGCCGGGUGCGCCAUUC ....(((((((((...))))....((.(((((((.......))))))).))(((((.......))))))))))......... (-31.75 = -31.75 + 0.00)

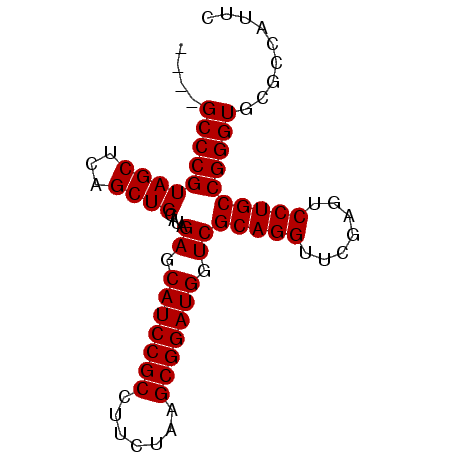
| Location | 1,947,318 – 1,947,400 |
|---|---|
| Length | 82 |
| Sequences | 6 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 1947318 82 - 1 GAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUU .......((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).. ( -33.90) >pst_1 1 77 - 1 -AAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGC---- -.........(((((((((.......)))))((.((((((((.....)))))))).))...............)))).---- ( -29.50) >pau_1 1 82 - 1 GAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGCUU .......((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).)).. ( -33.90) >pfo_1 1 79 - 1 -AAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGCGC-- -......((.(((((((((.......)))))((.((((((((.....)))))))).))...............)))).))-- ( -32.70) >psp_1 1 78 - 1 GAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGC---- ..........(((((((((.......)))))((.((((((((.....)))))))).))...............)))).---- ( -29.50) >pss_1 1 78 - 1 GAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGC---- ..........(((((((((.......)))))((.((((((((.....)))))))).))...............)))).---- ( -29.50) >consensus GAAUGGCGCACCCGGCAGGACUCGAACCUGCGACCAUCCGCUUAGAAGGCGGAUGCUCUAUCCAGCUGAGCUACGGGC____ ..........(((((((((.......)))))((.((((((((.....)))))))).))...............))))..... (-29.50 = -29.50 + 0.00)

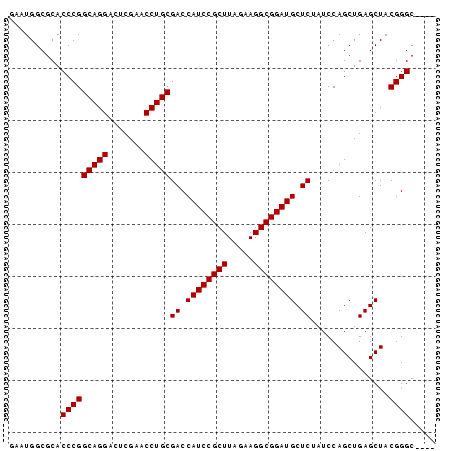
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:47:24 2007