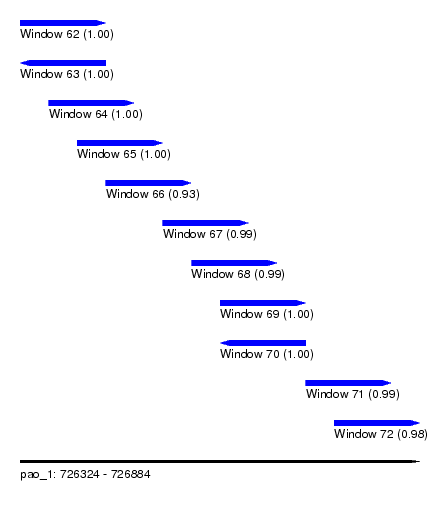
| Sequence ID | pao_1 |
|---|---|
| Location | 726,324 – 726,884 |
| Length | 560 |
| Max. P | 0.999744 |

| Location | 726,324 – 726,444 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -43.10 |
| Energy contribution | -42.97 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726324 120 + 1 GUGGGCAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....)))))))((...........))...))))))))))))))).... ( -45.40) >pel_1 2279 120 + 1 GUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... ( -42.30) >ppk_1 2279 120 + 1 GUGGGUAGUUUGACUGGGGCGGUCUCCUCCCAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... ( -42.30) >psp_1 2274 120 + 1 GUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... ( -42.80) >pss_1 2275 120 + 1 GUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... ( -42.80) >pst_1 2275 120 + 1 GUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... ( -42.80) >consensus GUGGGUAGUUUGACUGGGGCGGUCUCCUCCUAAAGAGUAACGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGA ....((((((.........((..(((((((...........)))))))..)).(((((((((..(((((((.....))))))).....((......))...))))))))))))))).... (-43.10 = -42.97 + -0.14)
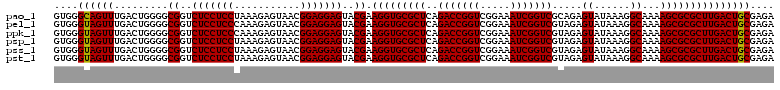
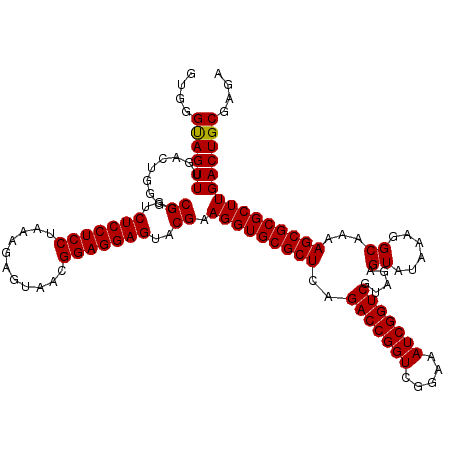
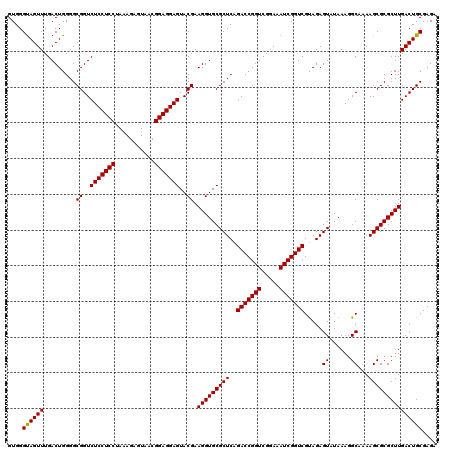
| Location | 726,324 – 726,444 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.50 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726324 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUGCGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUGCCCAC ....(((((...((((((((..((...........))(((((.........))))).)))))).....((..(((((((...........)))))))..)).....))...))))).... ( -36.60) >pel_1 2279 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. ( -31.10) >ppk_1 2279 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUGGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. ( -31.10) >psp_1 2274 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. ( -31.70) >pss_1 2275 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. ( -31.70) >pst_1 2275 120 - 1 UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. ( -31.70) >consensus UCUCGCAGUCAAGCGCGCUUUUGCCUUUAUACUCUACGACCGAUUUCCGACCGGUCUGAGCGCACCUUCGUACUCCUCCGUUACUCUUUAGGAGGAGACCGCCCCAGUCAAACUACCCAC ....((.(((..((((((....)).............(((((.........)))))...))))..........((((((...........))))))))).)).................. (-31.50 = -31.50 + -0.00)

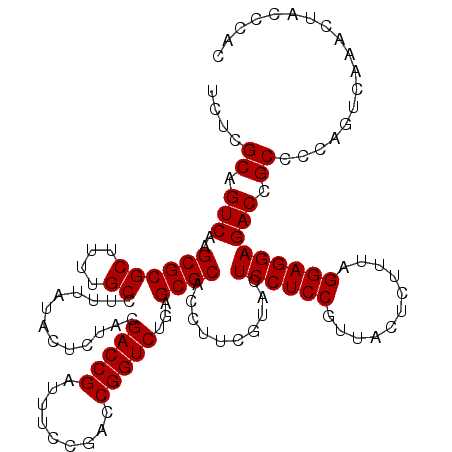
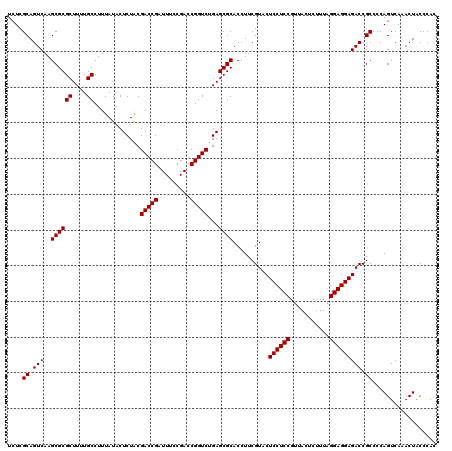
| Location | 726,364 – 726,484 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -40.40 |
| Energy contribution | -40.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.85 |
| SVM RNA-class probability | 0.999663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726364 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....)))))))((...........))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -41.00) >pel_1 2319 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -40.40) >ppk_1 2319 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -40.40) >psp_1 2314 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -40.40) >pss_1 2315 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -40.40) >pst_1 2315 120 + 1 CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) ( -40.40) >consensus CGGAGGAGUACGAAGGUGCGCUCAGACCGGUCGGAAAUCGGUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUC .......((((..(((((((((..(((((((.....))))))).....((......))...)))))))))..((((..(((......)))..)))))))).......((((.....)))) (-40.40 = -40.40 + -0.00)


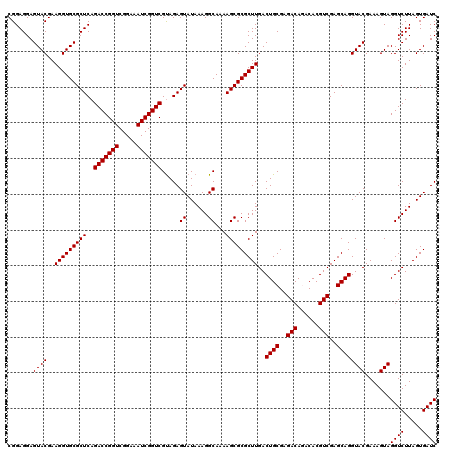
| Location | 726,404 – 726,524 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -39.27 |
| Energy contribution | -39.43 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726404 120 + 1 GUCGCAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((((......(((((......)).)))..))))))........(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))...... ( -39.90) >pel_1 2359 120 + 1 GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((..............((((..(((......)))..)))).(((....))).)))))........(((((((((.......))))))))))))).))))..... ( -39.80) >ppk_1 2359 120 + 1 GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((..............((((..(((......)))..)))).(((....))).)))))........(((((((((.......))))))))))))).))))..... ( -39.80) >psp_1 2354 120 + 1 GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......))))))))))))).))))..... ( -40.60) >pss_1 2355 120 + 1 GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......))))))))))))).))))..... ( -40.60) >pst_1 2355 120 + 1 GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((........((((((((((......).)).)))))))...(((....))).)))))........(((((((((.......))))))))))))).))))..... ( -40.60) >consensus GUCGUAGAGUAUAAAGGCAAAAGCGCGCUUGACUGCGAGACAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAA .((((.((((...(((((..............((((..(((......)))..)))).(((....))).)))))........(((((((((.......))))))))))))).))))..... (-39.27 = -39.43 + 0.17)

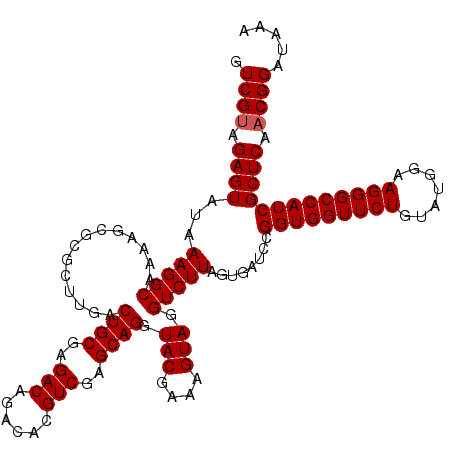
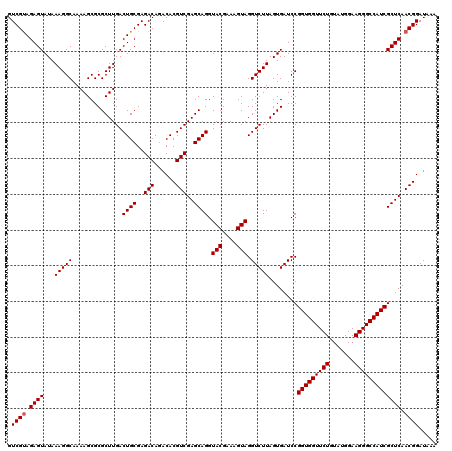
| Location | 726,444 – 726,564 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.56 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -38.30 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.00 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726444 120 + 1 CAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >pel_1 2399 120 + 1 CAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >ppk_1 2399 120 + 1 CAAACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >psp_1 2394 120 + 1 CAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >pss_1 2395 120 + 1 CAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >pst_1 2395 120 + 1 CAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). ( -38.30) >consensus CAGACACGUCGAGCAGGUACGAAAGUAGGUCUUAGUGAUCCGGUGGUUCUGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUU ......(((.((((...(((....)))((((.....)))).(((((((((.......))))))))))))).)))..........((((..(((......((......)).)))..)))). (-38.30 = -38.30 + 0.00)

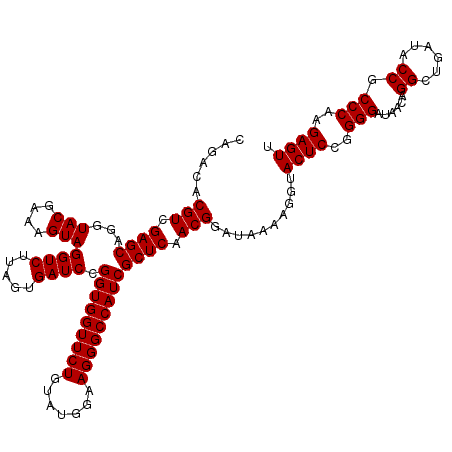
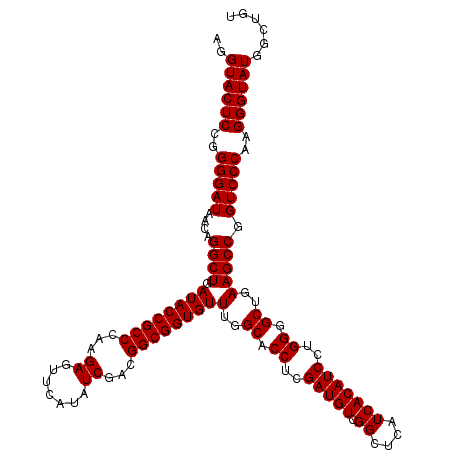
| Location | 726,524 – 726,644 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -47.70 |
| Energy contribution | -47.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726524 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >pfs_1 2478 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >ppk_1 2479 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >psp_1 2474 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >pss_1 2475 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >pst_1 2475 120 + 1 AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... ( -47.70) >consensus AGGUACUCCGGGGAUAACAGGCUGAUACCGCCCAAGAGUUCAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGU ..((((((..(((((....((((.((((((((...((.......))...))))))))(..((.((..(((((.((....)))))))..)).))..))))).)))))..))))))...... (-47.70 = -47.70 + -0.00)

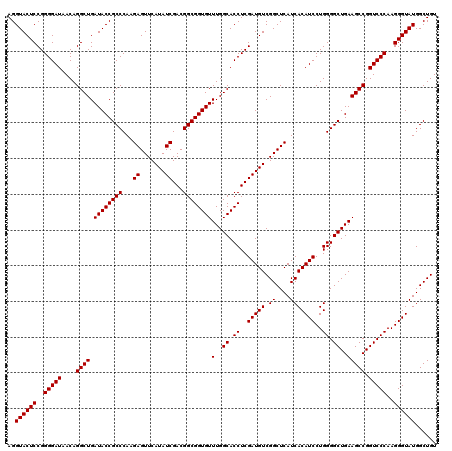
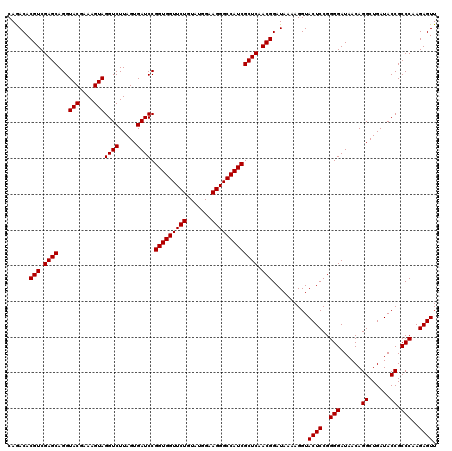
| Location | 726,564 – 726,684 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -46.80 |
| Energy contribution | -46.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726564 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >pfs_1 2518 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >ppk_1 2519 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >psp_1 2514 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >pss_1 2515 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >pst_1 2515 120 + 1 CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) ( -46.80) >consensus CAUAUCGACGGCGGUGUUUGGCACCUCGAUGUCGGCUCAUCACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUC ......(((((.((((.....)))).)....((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........)))) (-46.80 = -46.80 + 0.00)

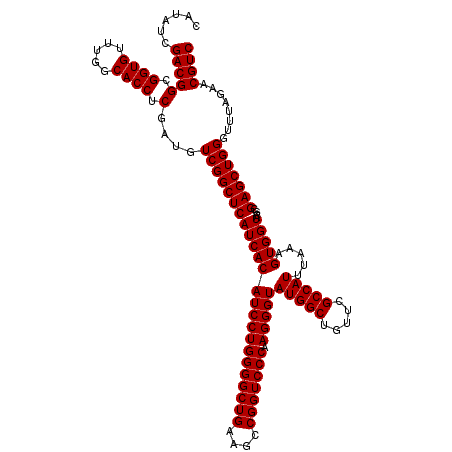
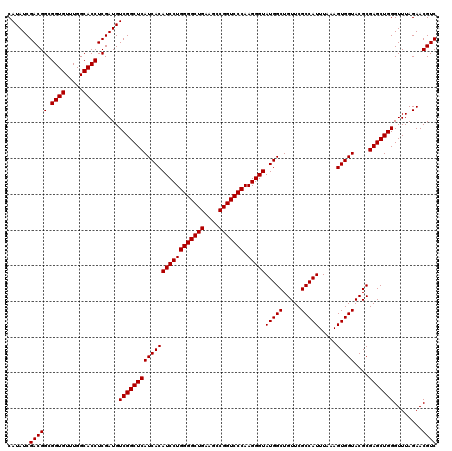
| Location | 726,604 – 726,724 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -48.90 |
| Consensus MFE | -45.90 |
| Energy contribution | -48.90 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726604 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >pfs_1 2558 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >ppk_1 2559 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >psp_1 2554 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >pss_1 2555 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >pst_1 2555 120 + 1 CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ ( -48.90) >consensus CACAUCCUGGGGCUGAAGCCGGUCCCAAGGGUAUGGCUGUUCGCCAUUUAAAGUGGUACGCGAGCUGGGUUUAGAACGUCGUGAGACAGUUCGGUCCCUAUCUGCCGUGGACGUUUGAGA (((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((.((.(((....(((((((....))).))))...))).....)))))).))........ (-45.90 = -48.90 + 3.00)
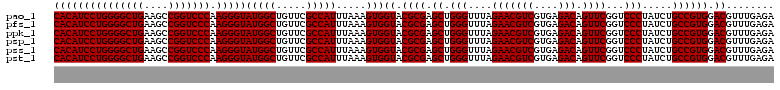
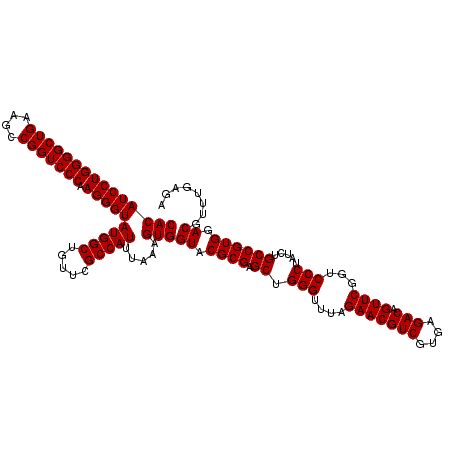
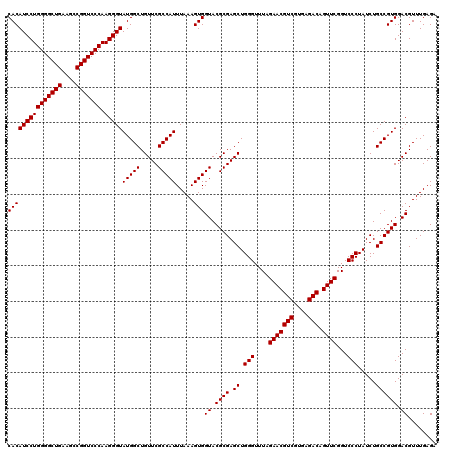
| Location | 726,604 – 726,724 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -38.50 |
| Energy contribution | -38.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726604 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >pfs_1 2558 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >ppk_1 2559 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >psp_1 2554 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >pss_1 2555 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >pst_1 2555 120 - 1 UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). ( -38.50) >consensus UCUCAAACGUCCACGGCAGAUAGGGACCGAACUGUCUCACGACGUUCUAAACCCAGCUCGCGUACCACUUUAAAUGGCGAACAGCCAUACCCUUGGGACCGGCUUCAGCCCCAGGAUGUG ......((((((..(((.....(((...((((.(((....)))))))....)))(((.((.((.(((......(((((.....))))).....))).)))))))...)))...)))))). (-38.50 = -38.50 + 0.00)

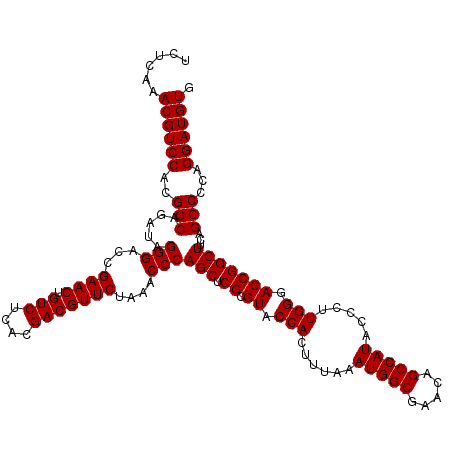
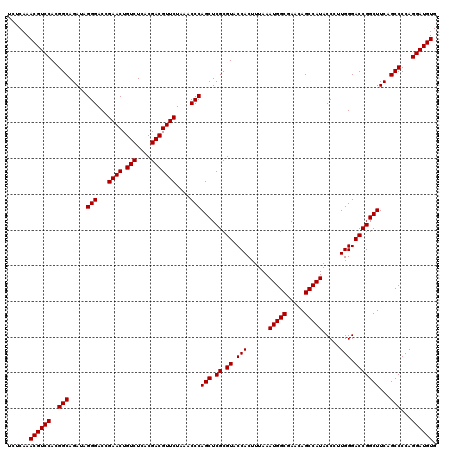
| Location | 726,724 – 726,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.11 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -43.90 |
| Energy contribution | -43.77 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726724 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAU ((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))..........((.....)).. ( -43.90) >pel_1 2679 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAU (((.((.((((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...(((....)))....)).)).))).... ( -46.00) >pfo_1 2677 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAU ...(((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...)))((.........))((.....)).. ( -43.40) >pfs_1 2678 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAUAGAUAACCGCUGAAAGCAU ((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))..........((.....)).. ( -43.90) >ppk_1 2679 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAU (((.((.((((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...(((....)))....)).)).))).... ( -46.00) >pst_1 2675 120 + 1 UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAU ((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))..........((.....)).. ( -43.90) >consensus UUUGAGAGGGGCUGCUCCUAGUACGAGAGGACCGGAGUGGACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAU ((((((...((((((((.......(.(((.(((((((.(......)))))))).))))(((((((((....))))))..)))))))))))...))))))..........((.....)).. (-43.90 = -43.77 + -0.14)

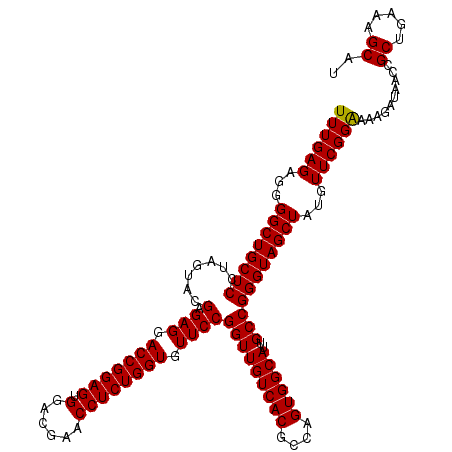
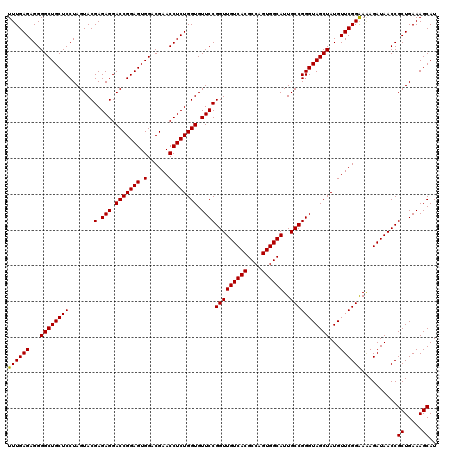
| Location | 726,764 – 726,884 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.39 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -38.87 |
| Energy contribution | -38.87 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 726764 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))..........(((.(((.(.(((((..((((....).)))....)))))...).))).))).. ( -39.00) >pel_1 2719 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))...(((((..(((((...........))))).........(((.....))))))))....... ( -41.50) >pf5_1 2716 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))..........(((.(((.(.(((((..((((....).)))....)))))...).))).))).. ( -38.80) >pfo_1 2717 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGGAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))((.(((.(..(((((...........)))))..).))).))((.((.(((....))))).)). ( -39.20) >pfs_1 2718 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAUAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUAGCCUCAAGAUGAGAUCUCACUGGAA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))..........(((.(((.(.(((((..((((....)).))....)))))...).))).))).. ( -38.80) >ppk_1 2719 120 + 1 ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAGAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGGA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))...(((((..(((((...........))))).........(((.....))))))))....... ( -41.50) >consensus ACGAACCUCUGGUGUUCCGGUUGUCACGCCAGUGGCAUUGCCGGGUAGCUAUGUUCGGAAAAGAUAACCGCUGAAAGCAUCUAAGCGGGAAACUUGCCUCAAGAUGAGAUCUCACUGGAA .(((((...((((..((((((((((((....))))))..))))))..)))).)))))..........(((.(((.(.(((((..((((....)).))....)))))...).))).))).. (-38.87 = -38.87 + 0.00)

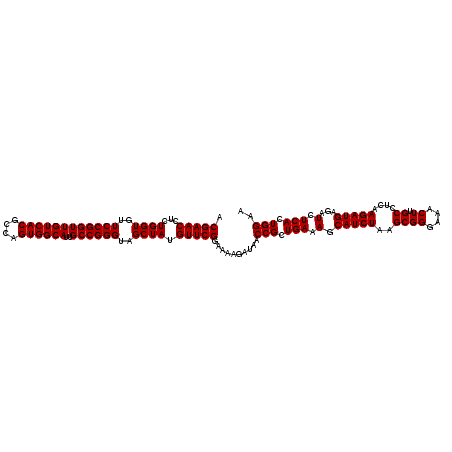
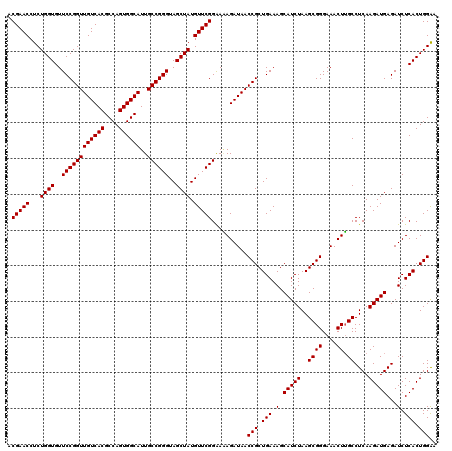
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:47:03 2007