
| Sequence ID | pao_1 |
|---|---|
| Location | 724,926 – 726,044 |
| Length | 1118 |
| Max. P | 0.999994 |

| Location | 724,926 – 725,046 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -41.62 |
| Energy contribution | -41.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.66 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.727199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724926 120 + 1 GCGCCUCAUGUAUCACUCUGGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -42.90) >pfs_1 880 120 + 1 GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -40.70) >ppk_1 880 120 + 1 GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -40.70) >psp_1 875 120 + 1 GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -40.70) >pss_1 875 120 + 1 GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -40.70) >pst_1 875 120 + 1 GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). ( -40.70) >consensus GCGCCUCAUGUAUCACUGUAGGGGGUAGAGCACUGUUUCGGCUAGGGGGUCAUCCCGACUUACCAAACCGAUGCAAACUCCGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGG .((((.(((((..(((((((((.....(((...(((.((((...(((((((.....))))).))...)))).)))..)))......))))).))))....)))))(....)....)))). (-41.62 = -41.07 + -0.55)

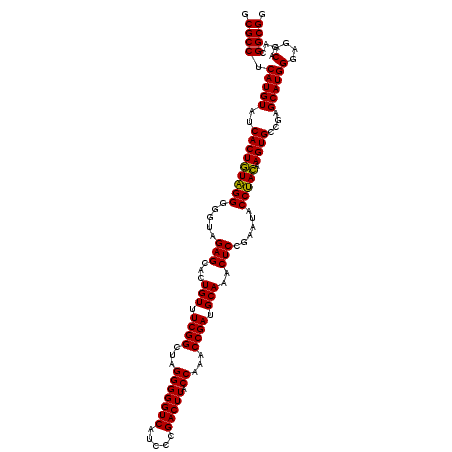
| Location | 725,006 – 725,126 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -38.81 |
| Energy contribution | -39.33 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725006 120 + 1 CGAAUACCCAGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAU ((..(((((....((((...))))(((.....((((((((..(....)..)))))))).....(....)..))).((((((.((((..(....)..))))))))))..).))))..)).. ( -39.00) >pf5_1 958 120 + 1 CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAU ......(((((....(((((((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))).))))...)))))....... ( -41.00) >pfo_1 960 120 + 1 CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAU ......(((((....(((((((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))).))))...)))))....... ( -41.00) >pfs_1 960 120 + 1 CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAU ......(((((....(((((((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))).))))...)))))....... ( -41.00) >pss_1 955 120 + 1 CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAU ......(((((....((((.((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))..))))...)))))....... ( -39.30) >pst_1 955 120 + 1 CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAU ......(((((....((((.((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))..))))...)))))....... ( -39.30) >consensus CGAAUACCUACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAU ......(((((....(((((((.((((.....((((((((..(....)..)))))))).....(....)..))))((((.........)))).....))).))))...)))))....... (-38.81 = -39.33 + 0.53)

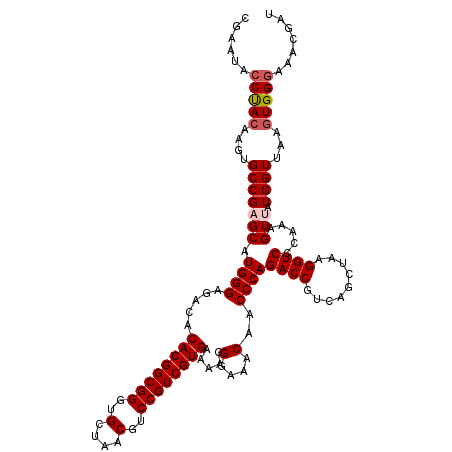
| Location | 725,046 – 725,166 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -35.96 |
| Energy contribution | -36.18 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725046 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((.(....)(((......))).))).((...(((((.(((((..(((((.............))))).)))))...)))))...))..))).))))))......... ( -36.72) >pf5_1 998 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..))))).......((((.....)))).))).))))))......... ( -37.40) >pfo_1 1000 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..))))).......((((.....)))).))).))))))......... ( -37.40) >pfs_1 1000 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..))))).......((((.....)))).))).))))))......... ( -37.40) >pss_1 995 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((.(....)(((......))).))).(((((((....(((((....(((.(((........))).)))))))).))))..))).....))).))))))......... ( -36.30) >pst_1 995 120 + 1 GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((.(....)(((......))).))).(((((((....(((((....(((.(((........))).)))))))).))))..))).....))).))))))......... ( -36.30) >consensus GUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCA ..((((((.((((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..))))).......((((.....)))).))).))))))......... (-35.96 = -36.18 + 0.22)

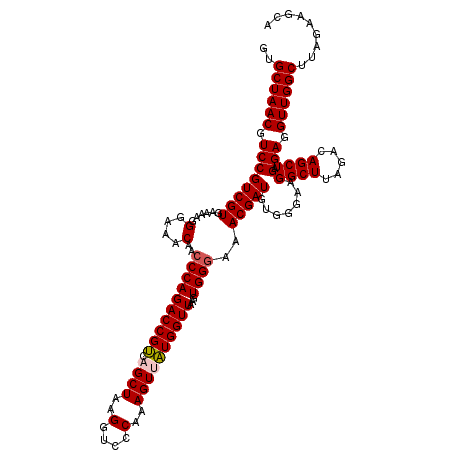
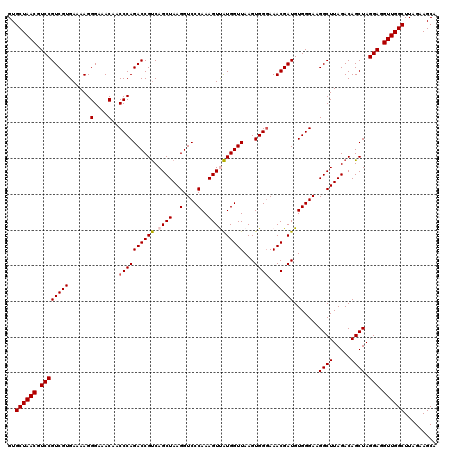
| Location | 725,046 – 725,166 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -27.22 |
| Consensus MFE | -26.45 |
| Energy contribution | -26.32 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725046 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC .........((.(((.(((..((((.(((((...(((((....(((........)))........))))).))))).))))((...(((......)))...)).....))).))).)).. ( -26.70) >pf5_1 998 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC .........((.(((.(((..(((.....)))...((((.((((((..........(((......)))..........)))))).))))...................))).))).)).. ( -26.55) >pfo_1 1000 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC .........((.(((.(((..(((.....)))...((((.((((((..........(((......)))..........)))))).))))...................))).))).)).. ( -26.55) >pfs_1 1000 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC .........((.(((.(((..(((.....)))...((((.((((((..........(((......)))..........)))))).))))...................))).))).)).. ( -26.55) >pss_1 995 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC ((((......((.....((..((((((..(((..((((((((.(((........))).)))....))))).....)))))))))..))(((((..........))))))).....)))). ( -28.50) >pst_1 995 120 - 1 UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC ((((......((.....((..((((((..(((..((((((((.(((........))).)))....))))).....)))))))))..))(((((..........))))))).....)))). ( -28.50) >consensus UGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGACGUUAGCAC .........((.(((.(((..(((.....)))...((((.((((((..........(((......)))..........)))))).))))...................))).))).)).. (-26.45 = -26.32 + -0.14)

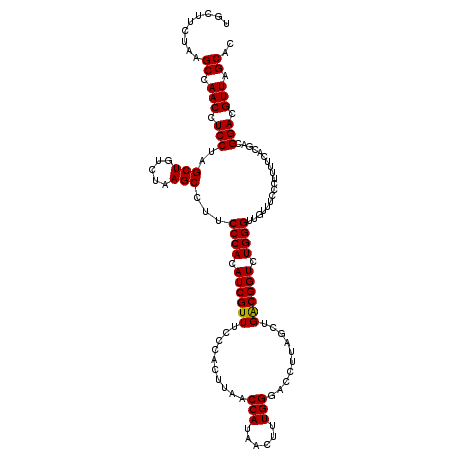
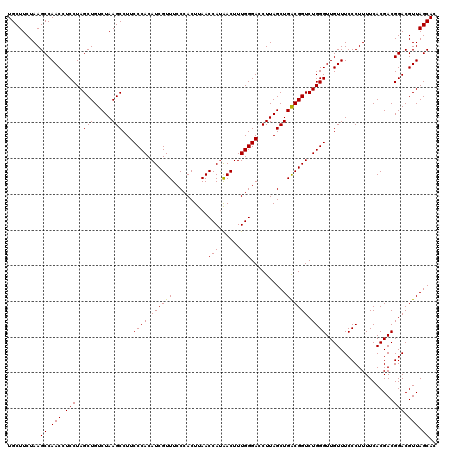
| Location | 725,086 – 725,206 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -32.66 |
| Consensus MFE | -32.33 |
| Energy contribution | -31.97 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725086 120 + 1 CCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((..(((((.............))))).)))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -32.82) >pf5_1 1038 120 + 1 UCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((.............((.(....).)).)))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -32.24) >pfo_1 1040 120 + 1 UCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((.............((.(....).)).)))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -32.24) >pfs_1 1040 120 + 1 UCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((.............((.(....).)).)))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -32.24) >pss_1 1035 120 + 1 UCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((....(((.(((........))).))))))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -33.20) >pst_1 1035 120 + 1 UCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((....(((.(((........))).))))))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... ( -33.20) >consensus UCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGU ....(((((.(((((....(((.(((........))).))))))))...))))).(.(((((((((.(((((.......))))).)))).......(((......)))..))))).)... (-32.33 = -31.97 + -0.36)

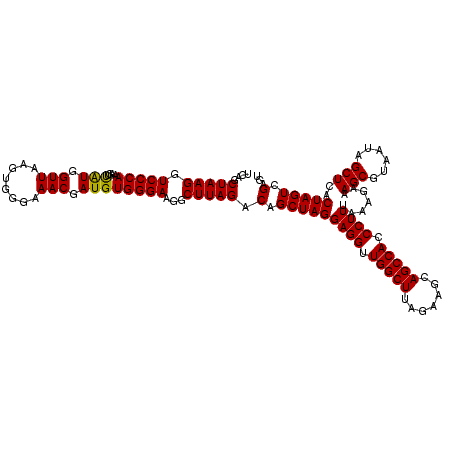
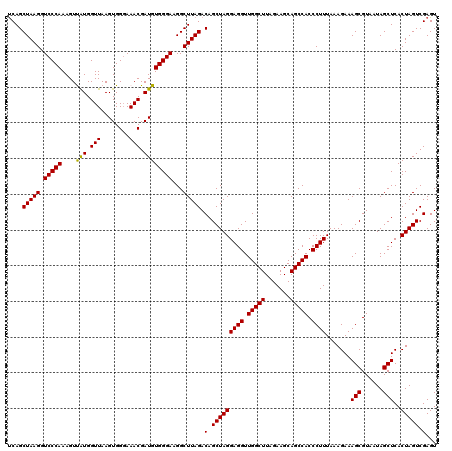
| Location | 725,126 – 725,246 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.28 |
| Mean single sequence MFE | -36.68 |
| Consensus MFE | -36.82 |
| Energy contribution | -36.68 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725126 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -38.60) >pf5_1 1078 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -36.30) >pfo_1 1080 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -36.30) >pfs_1 1080 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -36.30) >pss_1 1075 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -36.30) >pst_1 1075 120 + 1 GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... ( -36.30) >consensus GUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACAC ((((...(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))(((((((((....).)))....)))))....)))).... (-36.82 = -36.68 + -0.14)

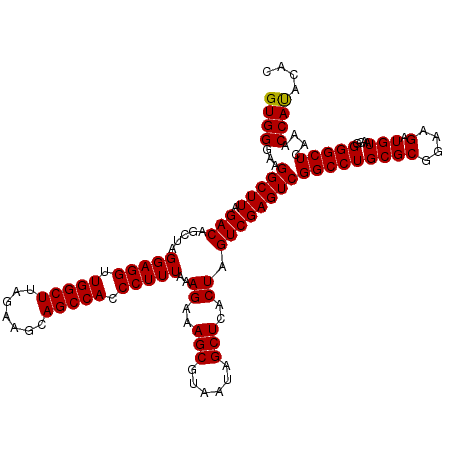
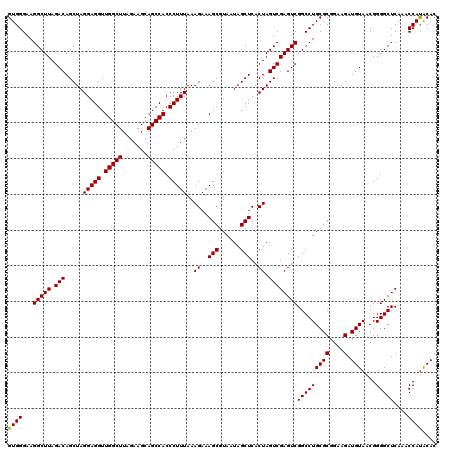
| Location | 725,206 – 725,324 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -40.94 |
| Energy contribution | -40.55 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725206 118 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..)))......((..((((....))))..)) ( -44.10) >pfo_1 1160 118 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..)))......((..((((....))))..)) ( -43.40) >ppk_1 1160 120 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUACGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..)))......((..((((....))))..)) ( -40.90) >psp_1 1155 120 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..)))......((..((((....))))..)) ( -41.50) >pss_1 1155 120 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..)))......((..((((....))))..)) ( -41.30) >pst_1 1155 120 + 1 CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..)))......((..((((....))))..)) ( -41.50) >consensus CGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUU_AGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCU .(((.(((((....).))))(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..)))......((..((((....))))..)) (-40.94 = -40.55 + -0.39)

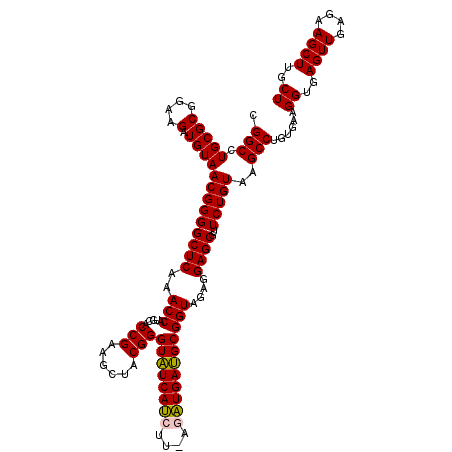
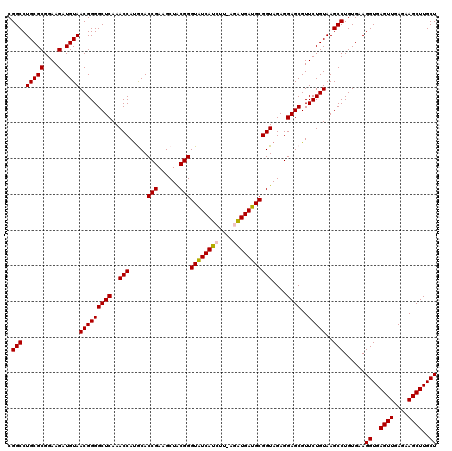
| Location | 725,246 – 725,364 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.27 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -35.62 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725246 118 + 1 CGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((...((((..(((((((..--..))))))).))))....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...((((......)))).))... ( -38.30) >pfo_1 1200 118 + 1 CGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...((((((((..--..)))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... ( -38.30) >ppk_1 1200 120 + 1 CGAAGCUACGGGUAUCAUCUUACGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...(((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... ( -35.80) >psp_1 1195 120 + 1 CGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...(((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... ( -36.40) >pss_1 1195 120 + 1 CGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...(((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... ( -36.20) >pst_1 1195 120 + 1 CGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...(((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... ( -36.40) >consensus CGAAGCUACGGGUAUCAUCUU_AGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACA ((..(((...(((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..)...)))..))... (-35.62 = -35.23 + -0.39)

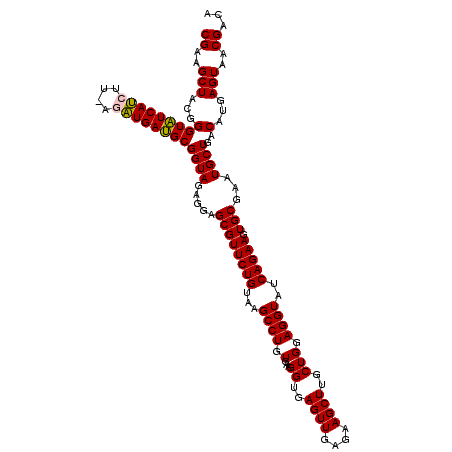
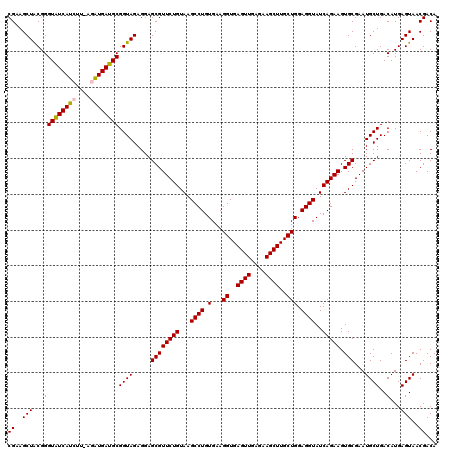
| Location | 725,284 – 725,404 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -35.39 |
| Energy contribution | -34.50 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.16 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725284 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUC ((((((((...((((.(...((..((((....))))..))).))))..))))).)))((((.((.................(((((((.....)))))))...(....)....)).)))) ( -36.70) >pel_1 1240 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUC ...(((((...((((.(...((..((((....))))..))).))))..)))))(.(((..((((......)))).......(((((((.....)))))))))))...((((....)))). ( -34.50) >ppk_1 1240 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUC ...(((((...((((.(...((..((((....))))..))).))))..)))))(.(((..((((......)))).......(((((((.....)))))))))))...((((....)))). ( -34.50) >psp_1 1235 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUC ...(((((...((((.(...((..((((....))))..))).))))..)))))(.(((..((((......)))).......(((((((.....)))))))))))...((((....)))). ( -35.10) >pss_1 1235 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUC ...(((((...((((.(...((..((((....))))..))).))))..)))))(.(((..((((......)))).......(((((((.....)))))))))))...((((....)))). ( -35.10) >pst_1 1235 120 + 1 GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUC ...(((((...((((.(...((..((((....))))..))).))))..)))))(.(((..((((......)))).......(((((((.....)))))))))))...((((....)))). ( -35.10) >consensus GCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUC ((.(((((...((((.(...((..((((....))))..))).))))..)))))))(((..((((......)))).......(((((((.....))))))))))(....)........... (-35.39 = -34.50 + -0.89)
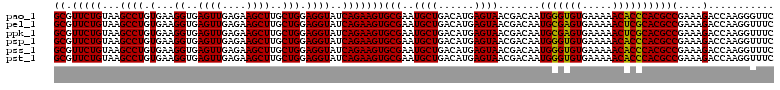
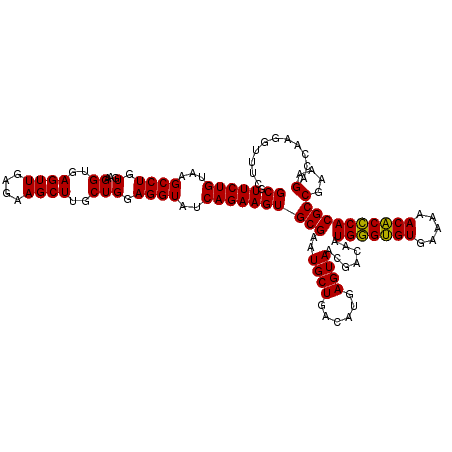
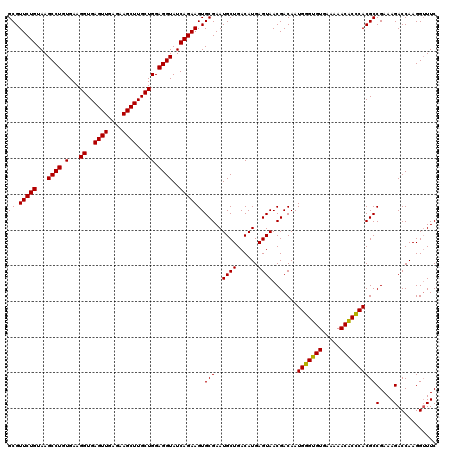
| Location | 725,324 – 725,444 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -35.39 |
| Energy contribution | -34.53 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725324 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAA (((.((((.....))))....((((((.((.((........(((((((.....)))))))...(....))))).....(((((((...((.....)).)))))))...)))))))))... ( -35.10) >pel_1 1280 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAA ((.(((((..(.....)..)))))(((.(((.(((((((..(((((((.....)))))))...(....)......))).((((((...((.....)).)))))))))).))))))))... ( -35.50) >ppk_1 1280 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAA ((.(((((..(.....)..)))))(((.(((.(((((((..(((((((.....)))))))...(....)......))).((((((...((.....)).)))))))))).))))))))... ( -35.50) >psp_1 1275 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAA ((.(((.(((..(((......)))....)))....((((..(((((((.....))))))).(((..........))).(((((((...((.....)).)))))))...)))))))))... ( -37.70) >pss_1 1275 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAA ((.(((((..(.....)..)))))(((.(((.(((((((..(((((((.....)))))))...(....)......))).((((((...((.....)).)))))))))).))))))))... ( -36.10) >pst_1 1275 120 + 1 GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAA ((.(((((..(.....)..)))))(((.(((.(((((((..(((((((.....)))))))...(....)......))).((((((...((.....)).)))))))))).))))))))... ( -36.10) >consensus GGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAA ..(((........(.(((..((((......)))).......(((((((.....)))))))))))....((((......(((((((...((.....)).)))))))......))))))).. (-35.39 = -34.53 + -0.86)
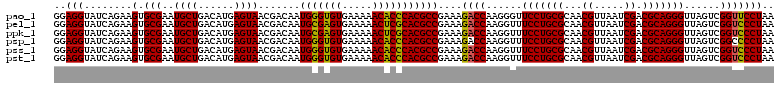
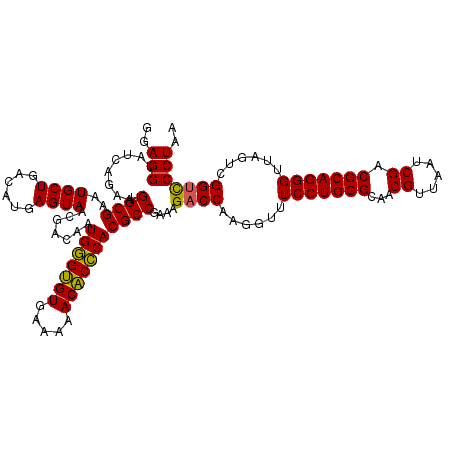
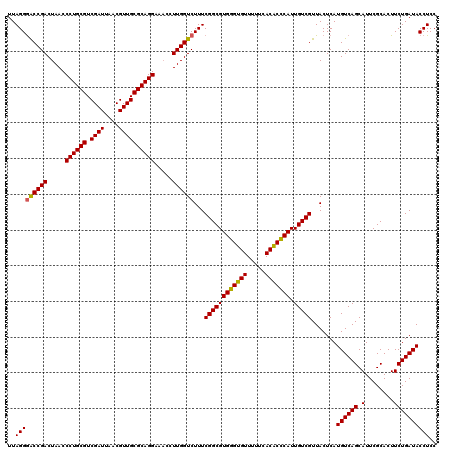
| Location | 725,324 – 725,444 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -40.43 |
| Consensus MFE | -41.14 |
| Energy contribution | -40.28 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.80 |
| Structure conservation index | 1.02 |
| SVM decision value | 5.79 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725324 120 - 1 UUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((....((((((.((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -37.90) >pel_1 1280 120 - 1 UUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -40.70) >ppk_1 1280 120 - 1 UUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -40.70) >psp_1 1275 120 - 1 UUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -40.70) >pss_1 1275 120 - 1 UUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -41.30) >pst_1 1275 120 - 1 UUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. ( -41.30) >consensus UUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCC ..(((((((((.....((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).))))).......((((((.(........)))))))))).. (-41.14 = -40.28 + -0.86)
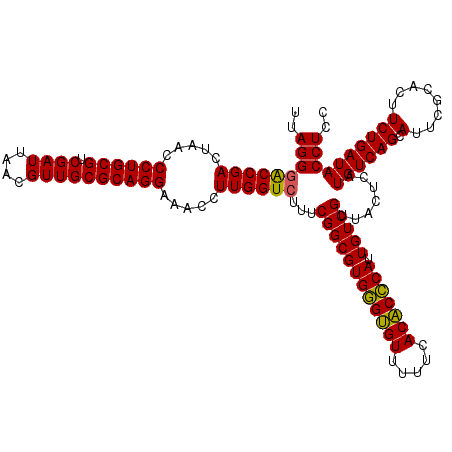
| Location | 725,364 – 725,484 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.11 |
| Mean single sequence MFE | -42.17 |
| Consensus MFE | -40.12 |
| Energy contribution | -40.77 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.60 |
| SVM RNA-class probability | 0.999927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725364 120 + 1 AUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAU .(((((((.....))))))).(((((..((((...))))((((((...((.....)).)))))).....)))))(((((.(((.(.(((....))).).)))..)))))........... ( -43.20) >pel_1 1320 120 + 1 AUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((....((((......(((((((...((.....)).)))))))......))))((...(((.(.(((....))).).)))...))......))).... ( -41.50) >ppk_1 1320 120 + 1 AUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((....((((......(((((((...((.....)).)))))))......))))((...(((.(.(((....))).).)))...))......))).... ( -41.50) >psp_1 1315 120 + 1 AUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((((.((((....))))((((((...((.....)).)))))).....))))).((...(((.(.(((....))).).)))...))............. ( -42.60) >pss_1 1315 120 + 1 AUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((....((((......(((((((...((.....)).)))))))......))))((...(((.(.(((....))).).)))...))......))).... ( -42.10) >pst_1 1315 120 + 1 AUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((....((((......(((((((...((.....)).)))))))......))))((...(((.(.(((....))).).)))...))......))).... ( -42.10) >consensus AUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAU .(((((((.....))))))).(((....((((......(((((((...((.....)).)))))))......))))((...(((.(.(((....))).).)))...))......))).... (-40.12 = -40.77 + 0.64)

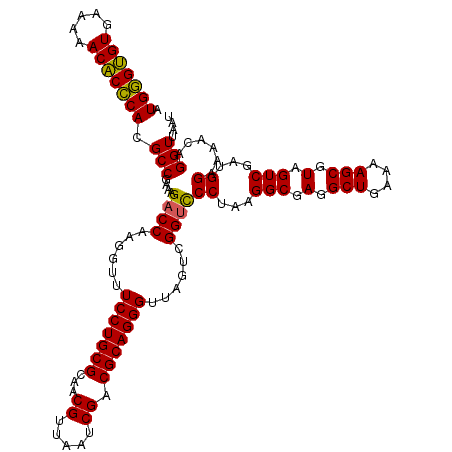
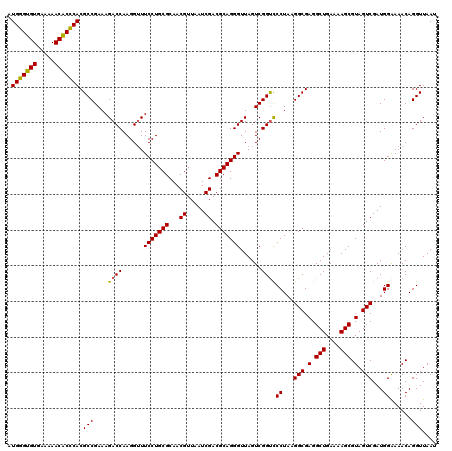
| Location | 725,364 – 725,484 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.11 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -39.81 |
| Energy contribution | -38.95 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.89 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.81 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725364 120 - 1 AUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAU ........................(((....)))..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))). ( -37.50) >pel_1 1320 120 - 1 AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAU ........................(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))). ( -39.40) >ppk_1 1320 120 - 1 AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAU ........................(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))). ( -39.40) >psp_1 1315 120 - 1 AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAU ......................(((((.....(((((......))))).((((((.((((((.((((....)))))))))).....))))))....)))))((((((.....)))))).. ( -40.10) >pss_1 1315 120 - 1 AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAU ........................(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))). ( -40.00) >pst_1 1315 120 - 1 AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAU ........................(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))). ( -40.00) >consensus AUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAU ........................(((....)))..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))). (-39.81 = -38.95 + -0.86)

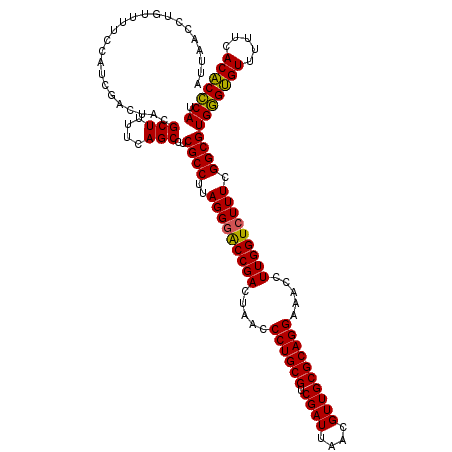
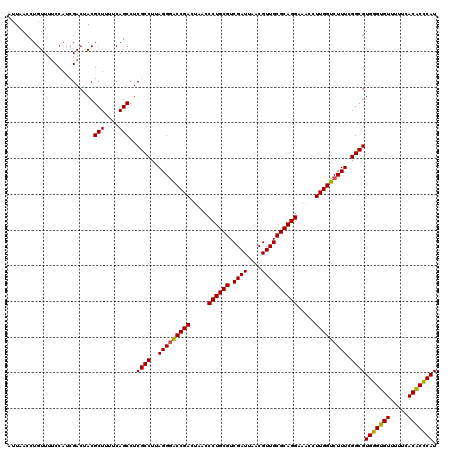
| Location | 725,404 – 725,524 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -38.95 |
| Consensus MFE | -37.49 |
| Energy contribution | -38.27 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725404 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....)))))).))... ( -40.00) >pel_1 1360 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))... ( -41.40) >pf5_1 1356 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))... ( -39.10) >ppk_1 1360 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))... ( -41.40) >psp_1 1355 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAA ........((((..((.(((((((((......))))....(((.(.(((....))).).)))...((((.(((((........)))))..))))......)))).).))..))))..... ( -34.00) >pst_1 1355 120 + 1 CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))... ( -37.80) >consensus CUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAA (((((...((.....)).)))))......((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))... (-37.49 = -38.27 + 0.78)

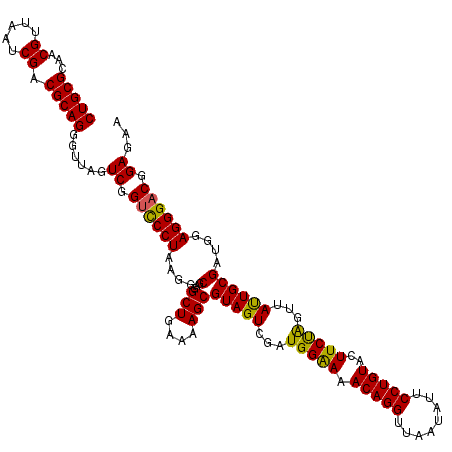
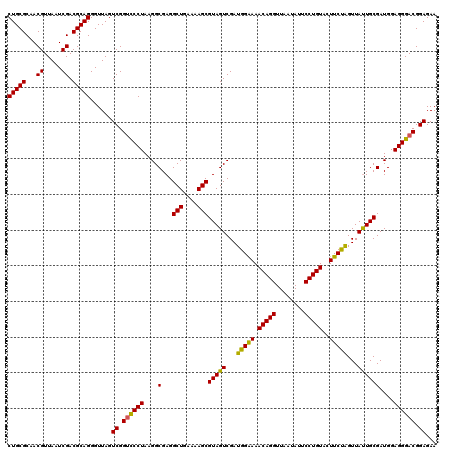
| Location | 725,404 – 725,524 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -27.54 |
| Energy contribution | -27.73 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725404 120 - 1 UUCUCCGUCCCUCCAUCGCAGUAACCAGAAGUACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG .................((.(((((..(((..(((((........))))))))..((((((...(((....)))...((.((((.......))))....))))))))....))))))).. ( -23.40) >pel_1 1360 120 - 1 UUCUCCGUCCCUCCAUCGCAAUAACUGGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ...((.((((((.....((......((((((.(((((........)))))))))))........(((....)))...))...)))))).))......(((((.((((....))))))))) ( -34.00) >pf5_1 1356 120 - 1 UUCUCCGUCCCUCCAUCGCAAUAACUAGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ...((.((((((.....((........((((.(((((........)))))))))..........(((....)))...))...)))))).))......(((((.((((....))))))))) ( -29.40) >ppk_1 1360 120 - 1 UUCUCCGUCCCUCCAUCGCAAUAACUGGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ...((.((((((.....((......((((((.(((((........)))))))))))........(((....)))...))...)))))).))......(((((.((((....))))))))) ( -34.00) >psp_1 1355 120 - 1 UUCUCCGUCCCUCCAUCGCAAUAACCAGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ................((((((.....((((.(((((........))))))))).((((((...(((....)))...((...((((.........))))))))))))....))))))... ( -29.00) >pst_1 1355 120 - 1 UUCUCCGUCCCUCCAUCGCAAUAACCAGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ...((.((((((.....((........((((.(((((........)))))))))..........(((....)))...))...)))))).))......(((((.((((....))))))))) ( -29.40) >consensus UUCUCCGUCCCUCCAUCGCAAUAACCAGAAGUACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAG ...((.((((((.....((........((((.(((((........)))))))))..........(((....)))...))...)))))).))......(((((.((((....))))))))) (-27.54 = -27.73 + 0.19)

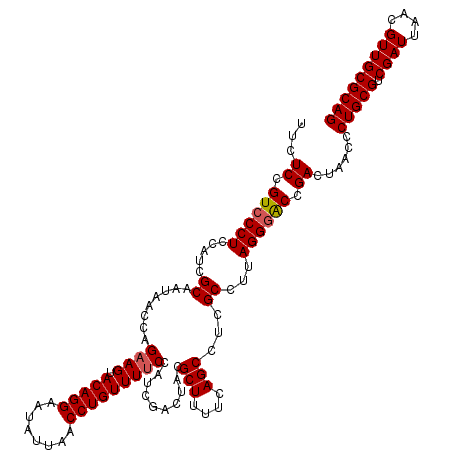
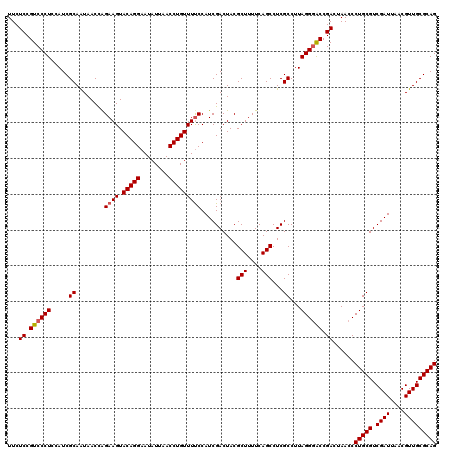
| Location | 725,444 – 725,564 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -38.35 |
| Energy contribution | -37.13 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.69 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725444 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUG ......(((....)))(((((.(((.(((((((((........)))))..)))).)))))))).......((((((..((.((((((....)))))).))..))))))............ ( -40.30) >pel_1 1400 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUG ......(((....)))(((((...(((((.(((((........)))))..)))))...))))).......((((((..((.((((((....)))))).))..))))))............ ( -38.80) >pf5_1 1396 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUG (((.(.(((....))).).)))........(((((........))))).((((..((....))...))))((((((..((.((((((....)))))).))..))))))............ ( -36.90) >ppk_1 1400 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUG ......(((....)))(((((...(((((.(((((........)))))..)))))...))))).......((((((..((.((((((....)))))).))..))))))............ ( -39.00) >pss_1 1395 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUG (((.(.(((....))).).)))........(((((........))))).(((((.((....)).).))))((((((..((.((((((....)))))).))..))))))............ ( -39.40) >pst_1 1395 120 + 1 GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUG .((.((((((....((.(((((........(((((........))))).(((((.((....)).).))))..........))))).)))))))).)).((((....)))).......... ( -36.80) >consensus GGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAAGUUUAAGGUG .((.(((((......((((((...(((((.(((((........)))))..)))))...))))))......((((((..((.((((((....)))))).))..)))))).)))))...)). (-38.35 = -37.13 + -1.22)

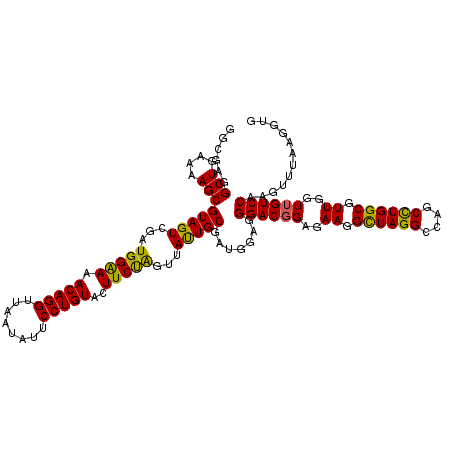
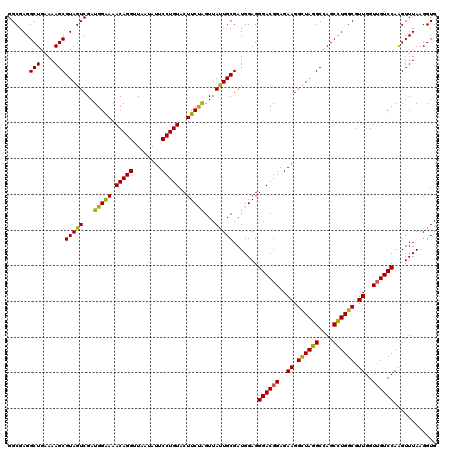
| Location | 725,484 – 725,604 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -40.97 |
| Energy contribution | -39.28 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725484 120 + 1 AUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCG .(((((((.(((((.((....)).).))))..)))).)))((((..((((.(((((..((((....))))..))))).))))....(((((((((.((.....))))))))))).)))). ( -39.20) >pel_1 1440 120 + 1 AUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCG .(((((((.((((((..........)))))).)))).)))((((..((((.(((((..((((....))))..))))).))))....((((((((..((....))..)))))))).)))). ( -45.40) >pf5_1 1436 120 + 1 AUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCG .(((((((.((((((..........)))))).)))).)))((((..((((.(((((..((((....))))..))))).))))....(((((((((.((.....))))))))))).)))). ( -40.30) >pfo_1 1438 120 + 1 AUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCG .(((((((.(((((.((....)).).))))..)))).)))((((..((((.(((((..((((....))))..))))).))))....(((((((((.((.....))))))))))).)))). ( -39.20) >ppk_1 1440 120 + 1 AUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCG .(((((((.((((((..........)))))).)))).))).....(((((((((((((.....)).))))))).............((((((((..((....))..)))))))).)))). ( -41.80) >pst_1 1435 120 + 1 AUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCG ...(((...(((((.((....)).).))))((((....((.((((((....)))))).))....)))))))............((((((((((((.((.....))))))))))..)))). ( -39.60) >consensus AUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUUUCAAGGCCG .(((((((.((((((..........)))))).)))).)))((((..((((.(((((..((((....))))..))))).))))....((((((((..((....))..)))))))).)))). (-40.97 = -39.28 + -1.69)

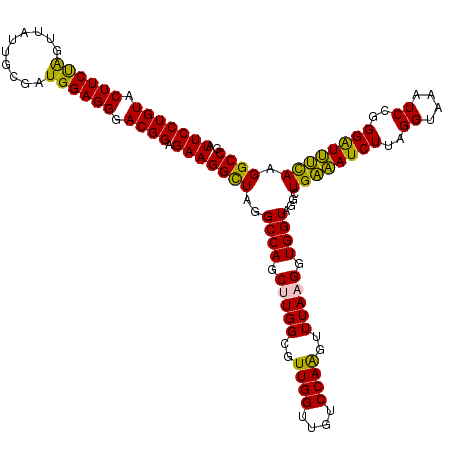
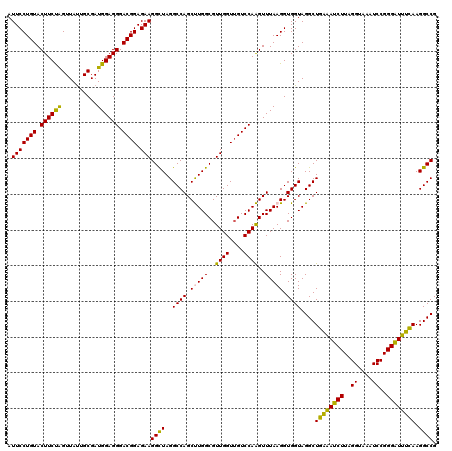
| Location | 725,524 – 725,644 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -43.52 |
| Consensus MFE | -46.59 |
| Energy contribution | -42.57 |
| Covariance contribution | -4.02 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.07 |
| SVM decision value | 5.84 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725524 120 + 1 GGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUU ((((..((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))...))))(((.((..((((((.....))))))...)).))) ( -42.30) >pel_1 1480 119 + 1 GGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUCAUU-AGAGCGCGAAGUGGUU ((((..((((.(((((..((((....))))..))))).)))).(((((((((((..((....))..)))))))..))))...))))(((.((..((((((...-.))))))...)).))) ( -47.00) >pf5_1 1476 120 + 1 GGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUU ((((..((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))...))))(((.((..((((((.....))))))...)).))) ( -43.20) >pfs_1 1478 120 + 1 GGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUUAACUUUUAGUUAACGAAGUGGUU ((((..((((.(((((..((((....))))..))))).)))).((((((((((((.((.....))))))))))..))))...))))(((.((..((((((.....))))))...)).))) ( -41.70) >ppk_1 1480 119 + 1 GGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCCUUU-AGGCGACGAAGUGGUU ((((.(((((((((((((.....)).))))))).............((((((((..((....))..)))))))).))))...))))(((.((..((((((...-.))))))...)).))) ( -43.60) >pst_1 1475 120 + 1 GGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACAUGAAGUGGUU ((((.((((((((((.((((((....))))........)).))))))((((((((.((.....))))))))))..))))...))))(((.((..((((((.....))))))...)).))) ( -43.30) >consensus GGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUUAUCUUUUAGAUGACGAAGUGGUU ((((.((((((((((.((((((....))))........)).))))))(((((((..((....))..)))))))..))))...))))(((.((..((((((.....))))))...)).))) (-46.59 = -42.57 + -4.02)

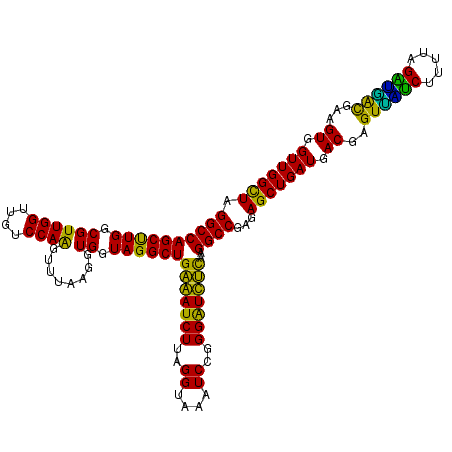
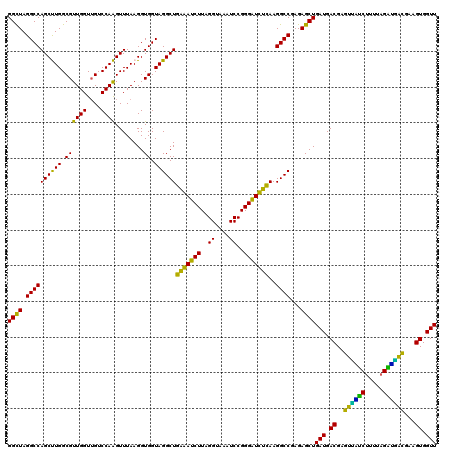
| Location | 725,524 – 725,644 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -22.86 |
| Energy contribution | -21.58 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725524 120 - 1 AACCACUUCGUCAUCUAAAAGACGACUCGUCAUCAGCUCUCGGCCUUGAAACCCCGGAUUUACCUAAGAUUUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCUGGCCUAGCC .......(((((........)))))..........(((...((((((((((.(..((.....))...).))))))..............(((((.(.......))))))..)))).))). ( -22.10) >pel_1 1480 119 - 1 AACCACUUCGCGCUCU-AAUGAGCACUCGUCAUCAGCUCUCGGCCUUGAGAUCCCGGAUUUGCCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCUGGCCUAGCC .........((((((.-...))))........((((((...(((.((((((((..((.....))...))))))))...............((((....)))).))).))))))....)). ( -31.20) >pf5_1 1476 120 - 1 AACCACUUCGUCACCUAAAAGGUAACUCGUCAUCAGCUCUCGGCCUUAGAACCCCGGAUUUACCUAAGAUUCCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCUGGCCUAGCC .........((.(((.....))).)).........(((...((((..........(((.((......)).)))................(((((.(.......))))))..)))).))). ( -20.70) >pfs_1 1478 120 - 1 AACCACUUCGUUAACUAAAAGUUAACUCGUCAUCAGCUCUCGGCCUUAAGAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCUGGCCUAGCC .........((((((.....)))))).........(((...((((...(((((..((.....))...))))).................(((((.(.......))))))..)))).))). ( -23.70) >ppk_1 1480 119 - 1 AACCACUUCGUCGCCU-AAAGGCAACUCGUCAUCAGCUCUCGGCCUUGAAAUCCCGGAUUUGCCUAAGAUUUCAGCCUACCACCUUAAACCUGGACAACCAACGCCAGGCUGGCCUAACC .........((.(((.-...))).))...............((((((((((((..((.....))...))))))))..............(((((.(.......))))))..))))..... ( -27.60) >pst_1 1475 120 - 1 AACCACUUCAUGUUCUAAAAGAACACUCGUCAUCAGCUCUCGGCCUUAAGAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACCUGGACUACCAACGCCAGGCUGGCCUAGCC ..........(((((.....)))))..........(((...((((...(((((..((.....))...))))).................(((((.(.......))))))..)))).))). ( -24.90) >consensus AACCACUUCGUCAUCUAAAAGACAACUCGUCAUCAGCUCUCGGCCUUAAAAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCUGGCCUAGCC .........((((((.....)))))).........(((...((((...(((((..((.....))...))))).................(((((.(.......))))))..)))).))). (-22.86 = -21.58 + -1.27)
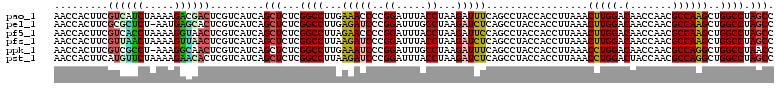
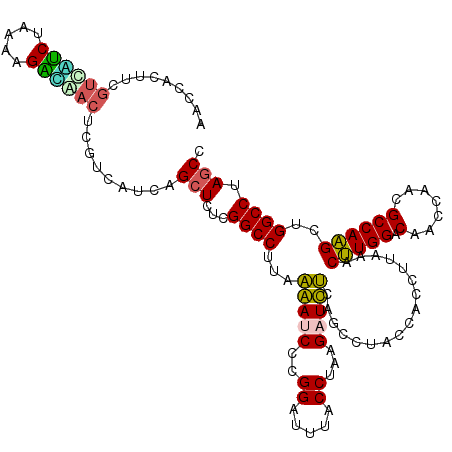
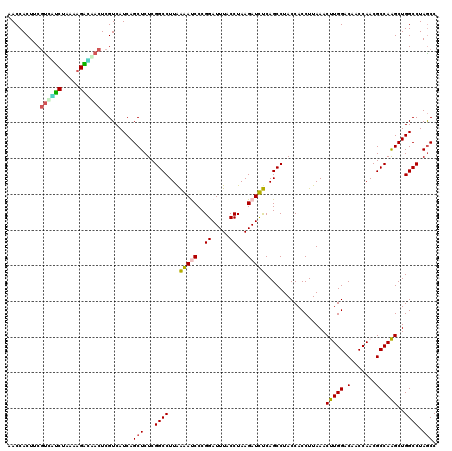
| Location | 725,564 – 725,684 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.49 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -39.34 |
| Energy contribution | -36.15 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.08 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725564 120 + 1 GUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAA ......(((((((((.((.....)))))))))))..(((..(((((...((.(.((((((.....))))))((((((((....)))..))))).........).))..)))))..))).. ( -35.00) >pel_1 1520 119 + 1 GUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUCAUU-AGAGCGCGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAA ...(((((((((((..((....))..)))))))..))))..(((((........((((((...-.))))))((((((((....)))..))))).......)))))............... ( -40.00) >pf5_1 1516 120 + 1 GUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAA ...((((((((((((.((.....))))))))))..))))(..(((.........((((((.....))))))...(((((....))))))))..).....(((((....)))))....... ( -35.80) >pfo_1 1518 119 + 1 GUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUUGCCUUU-AGGCGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAA ...((((((((((((.((.....))))))))))..))))(..(((.........((((((...-.))))))...(((((....))))))))..).....(((((....)))))....... ( -36.70) >pfs_1 1518 120 + 1 GUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUUAACUUUUAGUUAACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGGUAA ...((((((((((((.((.....))))))))))..))))(..(((.........((((((.....))))))...(((((....))))))))..).....(((((....)))))....... ( -34.30) >ppk_1 1520 119 + 1 GUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCCUUU-AGGCGACGAAGUGGUUGAUACCAUGCUUCCAAGAAAAGCUCCUAAGCUUCAGAUAA ...(((((((((((..((....))..)))))))..))))..(((((........((((((...-.))))))((((((((....)))..))))).......)))))............... ( -37.00) >consensus GUAGGCUGAAAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCCUUU_AGGUGACGAAGUGGUUGAUGCCAUGCUUCCAAGAAAAGCUUCUAAGCUUCAGAUAA ...(((((((((((..((....))..)))))))..))))(..(((.........((((((.....))))))...(((((....))))))))..).....(((((....)))))....... (-39.34 = -36.15 + -3.19)
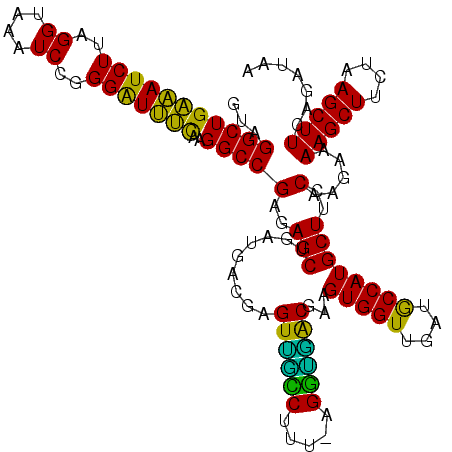
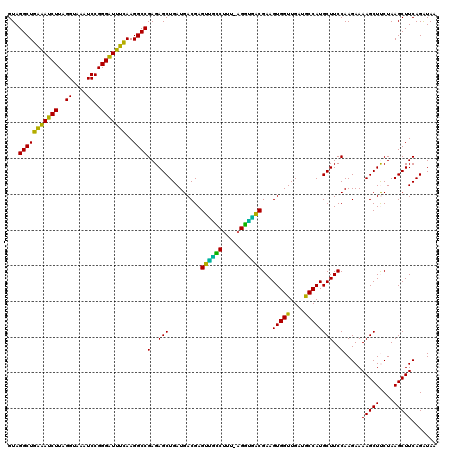
| Location | 725,684 – 725,804 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -33.33 |
| Energy contribution | -32.97 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725684 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCGACCACCUGUGUCGGUUUGGGGUACGGUUCCUGG (((((((..........((((..(((((........)))..))..))))....(((....)))..)))))...(((((((..(((.((((....).))))))..)))))))))....... ( -36.20) >pel_1 1639 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGGUACGGUUCCCAG ..(((((..........((((..(((((........)))..))..))))....(((....)))..))))).(((....((.(((((....(((......)))...))))).))...))). ( -33.10) >pf5_1 1636 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUUCCUAG ..(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((....((.(((((....(((......)))...))))).))..))... ( -34.00) >ppk_1 1639 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCUAACCACCUGUGUCGGUUUGGGGUACGGUUCCCAG ..(((((..........((((..(((((........)))..))..))))....(((....)))..))))).(((....((.(((((....(((......)))...))))).))...))). ( -33.10) >pss_1 1635 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUUCCUGG (((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((....((.(((((....(((......)))...))))).))..)).)) ( -34.40) >pst_1 1635 120 - 1 CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUUCCUGG (((((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((....((.(((((....(((......)))...))))).))..)).)) ( -34.40) >consensus CCGGCGCACCUUCUCCCGAAGUUACGGUGCCAUUUUGCCUAGUUCCUUCACCCGAGUUCUCUCAAGCGCCUUGGUAUUCUCUACCCAACCACCUGUGUCGGUUUGGGGUACGGUUCCUAG ..(((((..........((((..(((((........)))..))..))))....(((....)))..)))))..((....((.(((((((((((....)).))))..))))).))..))... (-33.33 = -32.97 + -0.36)

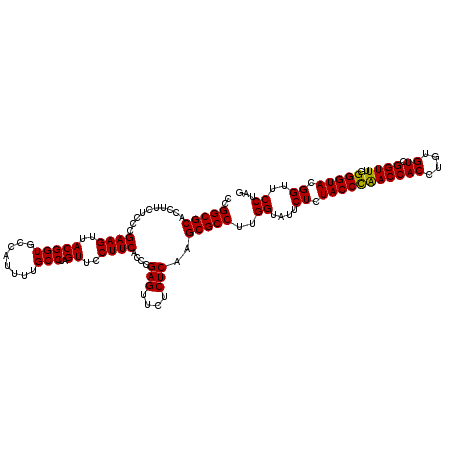
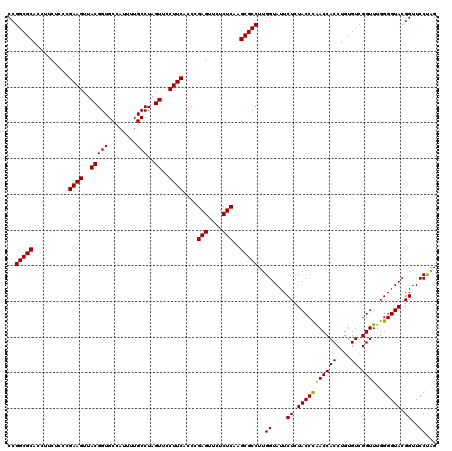
| Location | 725,724 – 725,844 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -41.33 |
| Energy contribution | -40.05 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725724 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))((((((((((((...(((.....)))....))))))))))))... ( -45.20) >pf5_1 1676 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))... ( -39.80) >pfs_1 1678 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))... ( -39.80) >ppk_1 1679 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))... ( -45.40) >pss_1 1675 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..)))))... ( -42.40) >pst_1 1675 120 + 1 AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGA ......((....((((((((....))))))))..))..............(((((....(....).....)))))(((((((.((((...(((.....)))....)))).)))))))... ( -45.10) >consensus AGAAUACCAAGGCGCUUGAGAGAACUCGGGUGAAGGAACUAGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUACUCCGUAAGCCCAUGCCGGUCGA ......((....((((((((....))))))))..))..............(((((.....(((....))))))))(((((((.((((...(((.....)))....)))).)))))))... (-41.33 = -40.05 + -1.28)

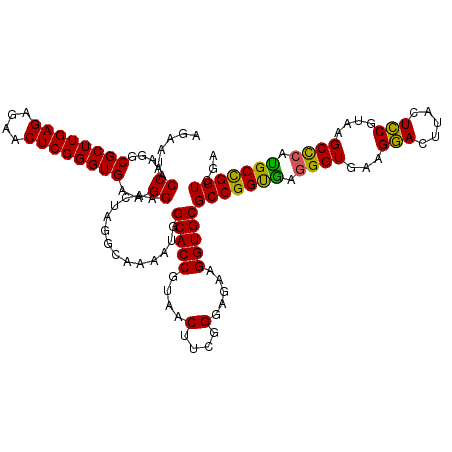
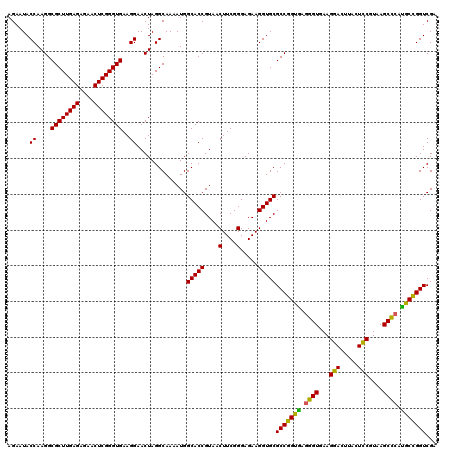
| Location | 725,764 – 725,884 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -44.45 |
| Consensus MFE | -43.72 |
| Energy contribution | -42.45 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.62 |
| SVM RNA-class probability | 0.999929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725764 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))((((((((((((...(((.....)))....)))))))))))).........)).)))((((....(((((.....))))))))). ( -46.70) >pf5_1 1716 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).)))((((....(((((.....))))))))). ( -41.30) >pfs_1 1718 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).)))((((....(((((.....))))))))). ( -41.30) >ppk_1 1719 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGCGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).)))((((....(((((.....))))))))). ( -46.90) >pss_1 1715 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))(((((..(.(((...(((.....)))....))).)..))))).........)).)))((((....(((((.....))))))))). ( -43.90) >pst_1 1715 120 + 1 AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((((((....(....).....)))))(((((((.((((...(((.....)))....)))).))))))).........)).)))((((....(((((.....))))))))). ( -46.60) >consensus AGGCAAAAUGGCACCGUAACUUCGGGAGAAGGUGCGCCGGUGAGGGUGAAGGACUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCA .(((....(((....(((.((((....)))).)))(((((((.((((...(((.....)))....)))).)))))))........))).)))((((....(((((.....))))))))). (-43.72 = -42.45 + -1.27)
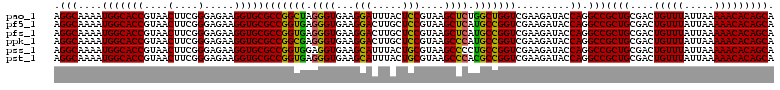
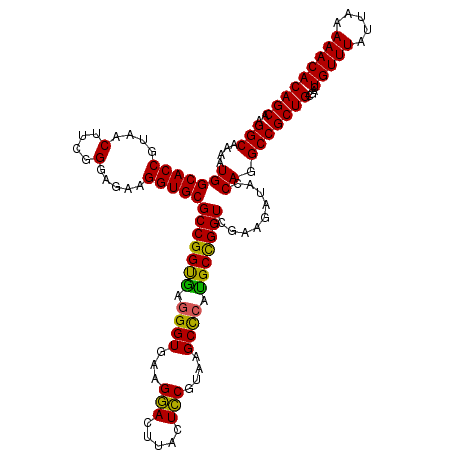
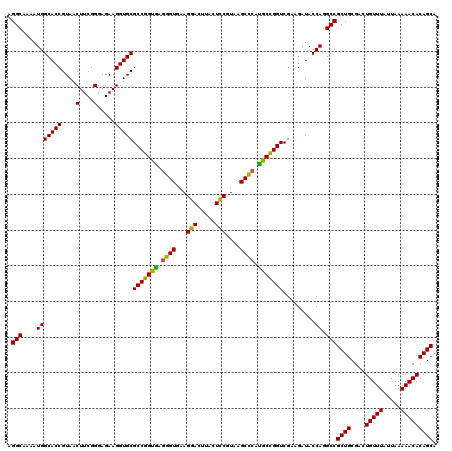
| Location | 725,804 – 725,924 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.06 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -35.56 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725804 120 + 1 CUAGGGUGAAGGAUUUACUCCGUAAGCUCUGGCUGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..(((.....)))....((..((((((((.......)))).))))..))...(((((.....)))))..((((((((....(((....)))......))))))))..))))) ( -36.00) >pf5_1 1756 120 + 1 UGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..(((.....)))......(((((((((((..........))))((((....(((((.....))))))))).((.(((...(((....)))...))).)))))))))))))) ( -36.70) >pfs_1 1758 120 + 1 UGAGGGUGAAGGACUUGCUCCGUAAGCUCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..(((.....)))......(((((((((((..........))))((((....(((((.....))))))))).((.(((...(((....)))...))).)))))))))))))) ( -36.70) >ppk_1 1759 120 + 1 CGAGGGUGAAGGACUUGCUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..((.(((((...)))))))((((((((((..........))))((((....(((((.....))))))))).((.(((...(((....)))...))).)))))))).))))) ( -38.10) >pss_1 1755 120 + 1 UGGAGGUGAAGCAUUUACUGCGUAAGCCCCUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..((((((..(((((..((....((((((.......))).))).((((....(((((.....))))))))).....))...(((....))).)))))..))))))..))))) ( -35.50) >pst_1 1755 120 + 1 UGAGGGUGAAGCAUUUACUGCGUAAGCCCACGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((...........((....)).((((((((((..........))))((((....(((((.....))))))))).((.(((...(((....)))...))).)))))))).))))) ( -38.70) >consensus UGAGGGUGAAGGACUUACUCCGUAAGCCCAUGCCGGUCGAAGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCU ...(((((..(((.....)))((((((...((((((((..........)))))..)))...))))))......((((.(.((.(((...(((....)))...))).))).)))).))))) (-35.56 = -34.90 + -0.66)

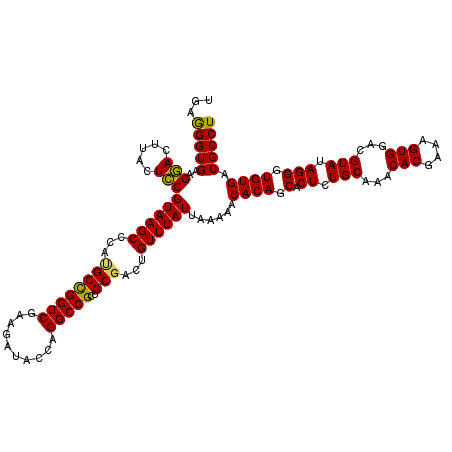
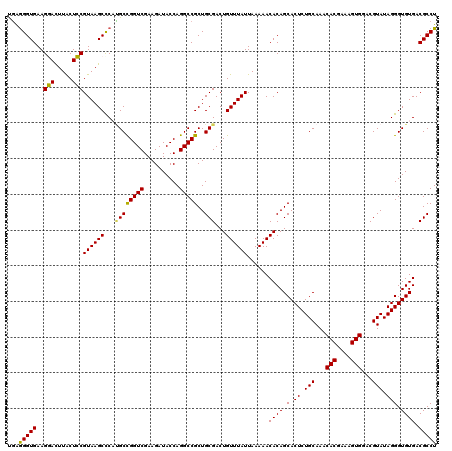
| Location | 725,844 – 725,964 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -37.55 |
| Energy contribution | -37.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725844 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).)).(((..((((.(((((....)))......)).))))..)))..... ( -39.10) >pf5_1 1796 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).))..((..((((.(((((....)))......)).))))..))...... ( -37.30) >pfo_1 1797 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).))..((..((((.(((((....)))......)).))))..))...... ( -37.30) >pfs_1 1798 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).))..((..((((.(((((....)))......)).))))..))...... ( -37.30) >pss_1 1795 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).)).(((..((((.(((((....)))......)).))))..)))..... ( -39.10) >pst_1 1795 120 + 1 AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).)).(((..((((.(((((....)))......)).))))..)))..... ( -39.10) >consensus AGAUACCAGGCCGCUGCGACUGUUUAUUAAAAACACAGCACUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGAAAGC ......((.(((((((....(((((.....))))))))).((.(((...(((....)))...))).))))).))..((..((((.(((((....)))......)).))))..))...... (-37.55 = -37.55 + 0.00)

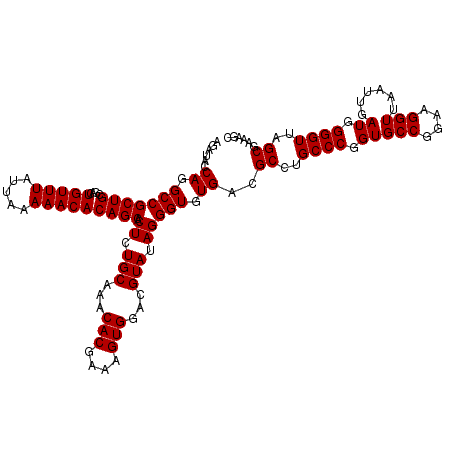
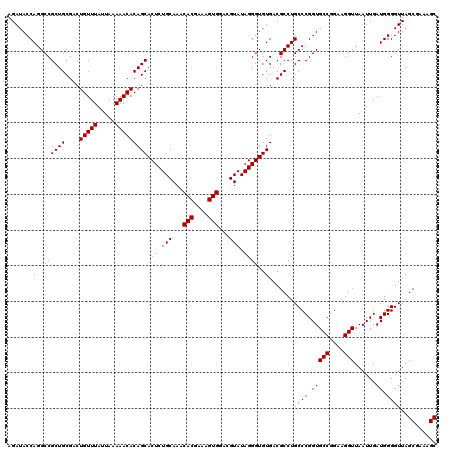
| Location | 725,884 – 726,004 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -44.40 |
| Energy contribution | -45.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725884 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -46.30) >pf5_1 1836 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -44.50) >pfo_1 1837 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -44.50) >pfs_1 1838 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -44.50) >pss_1 1835 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -46.30) >pst_1 1835 120 + 1 CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... ( -46.30) >consensus CUCUGCAAACACGAAAGUGGACGUAUAGGGUGUGACGCCUGCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGAAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAAC ((.(((...(((....)))...))).))......(((.(((((...((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...))))).)))... (-44.40 = -45.40 + 1.00)
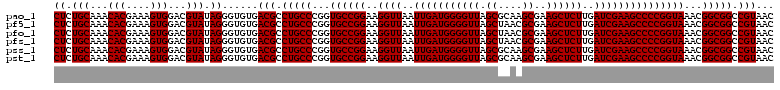

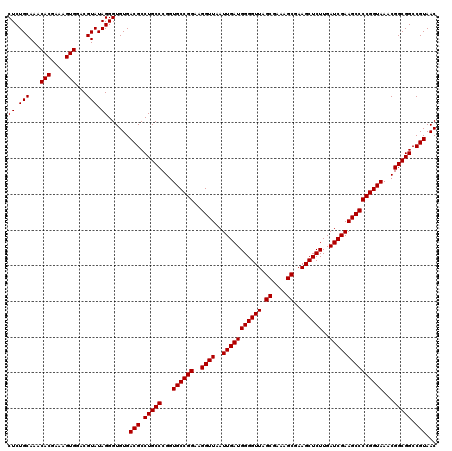
| Location | 725,924 – 726,044 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -42.40 |
| Energy contribution | -43.40 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 725924 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -44.30) >pf5_1 1876 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -42.50) >pfo_1 1877 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -42.50) >pfs_1 1878 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCUAACGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -42.50) >pss_1 1875 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -44.30) >pst_1 1875 120 + 1 GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGCAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... ( -44.30) >consensus GCCCGGUGCCGGAAGGUUAAUUGAUGGGGUUAGCGAAAGCGAAGCUCUUGAUCGAAGCCCCGGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAG ((((..((((((..((((..(((((((((((.((....))..))))))..)))))))))))))))...(((((((((.......))))))..((((..........)))))))))))... (-42.40 = -43.40 + 1.00)
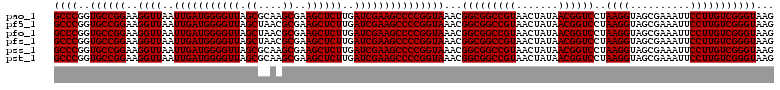
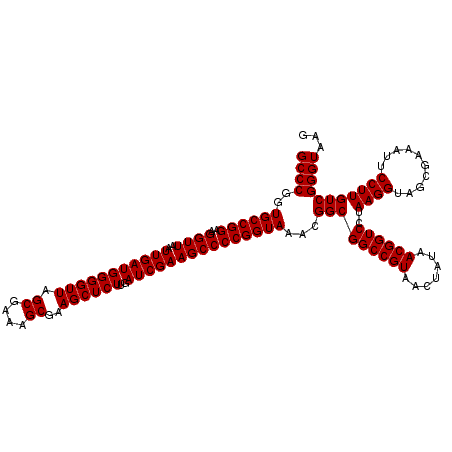
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:46:46 2007