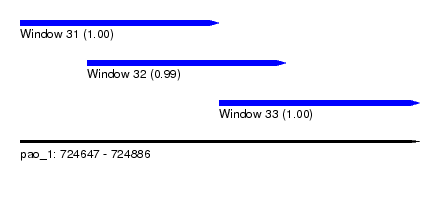
| Sequence ID | pao_1 |
|---|---|
| Location | 724,647 – 724,886 |
| Length | 239 |
| Max. P | 0.997320 |

| Location | 724,647 – 724,766 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -39.44 |
| Consensus MFE | -39.28 |
| Energy contribution | -38.78 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724647 119 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCC ......((((.....))))...(((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))))))) ( -38.81) >pel_1 601 120 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCC ......((((.....))))...(((((.......(((((((((((((..((((..((.....))..))))((.((....))..((((.....))))))....)))))))))))))))))) ( -39.81) >pf5_1 599 119 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCC ......((((.....))))...(((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))) ( -39.51) >ppk_1 601 119 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCC ......((((.....))))...(((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).-....)))))))))))))))))) ( -39.51) >pss_1 596 119 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCC ......((((.....))))...(((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))) ( -39.51) >pst_1 596 119 + 1 GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCC ......((((.....))))...(((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))) ( -39.51) >consensus GACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCC ......((((.....))))...(((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....))))......)))))))))))))))))) (-39.28 = -38.78 + -0.50)

| Location | 724,687 – 724,806 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.98 |
| Mean single sequence MFE | -41.55 |
| Consensus MFE | -39.10 |
| Energy contribution | -39.27 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724687 119 + 1 CAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGG (((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))((.(((((((((....))))...)))))))..................... ( -38.00) >pel_1 641 120 + 1 CAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGG .((((((..((((..((.....))..))))((.((....))..((((.....))))))....))))))((((....))))..((((...((((((..(....)..))))))..))))... ( -42.30) >pf5_1 639 119 + 1 CAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGG (((((((..((((..((.....))..))))...((....))((((((.....)))).-))..))))))).........((..((((...((((((..(....)..))))))..)))).)) ( -41.00) >ppk_1 641 119 + 1 CAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGG .((((((..((((..((.....))..))))...((....))..((((.....)))).-....))))))((((....))))..((((...((((((..(....)..))))))..))))... ( -42.00) >pss_1 636 119 + 1 CAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGG (((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))........((((..(((...((((((..(....)..))))))..))))))) ( -43.00) >pst_1 636 119 + 1 CAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGG (((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))........((((..(((...((((((..(....)..))))))..))))))) ( -43.00) >consensus CAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGG .((((((..((((..((.....))..))))...((....))..((((.....))))......))))))(((......)))..((((...((((((..(....)..))))))..))))... (-39.10 = -39.27 + 0.17)
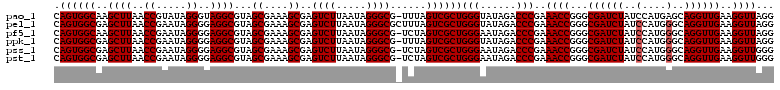
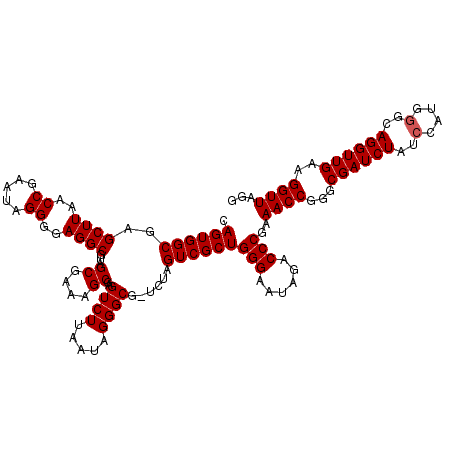
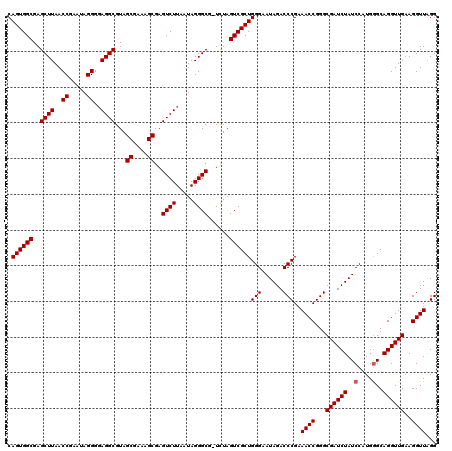
| Location | 724,766 – 724,886 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -36.37 |
| Energy contribution | -35.98 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724766 120 + 1 GAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).))).))...))........ ( -37.00) >pfs_1 720 120 + 1 GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ ( -37.30) >ppk_1 720 120 + 1 GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ ( -37.70) >psp_1 715 120 + 1 GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ ( -37.30) >pss_1 715 120 + 1 GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ ( -37.30) >pst_1 715 120 + 1 GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ ( -37.30) >consensus GAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAA ....((...((.((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).))...))........ (-36.37 = -35.98 + -0.39)

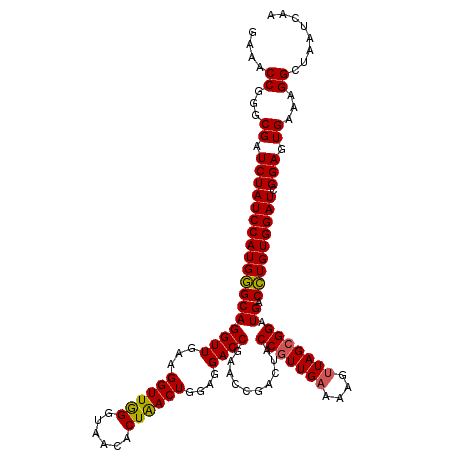
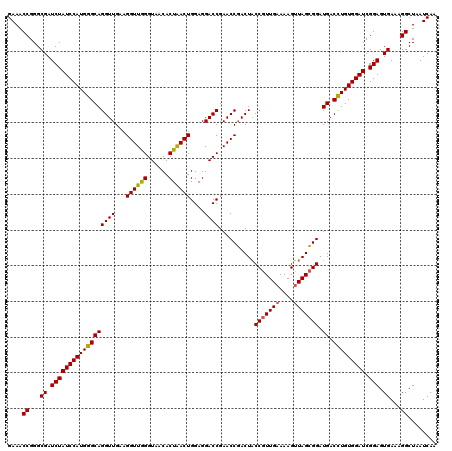
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:46:11 2007