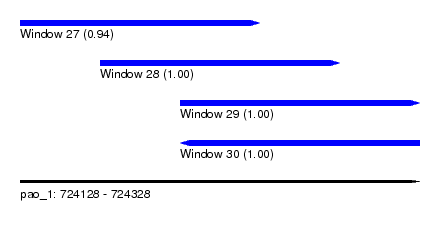
| Sequence ID | pao_1 |
|---|---|
| Location | 724,128 – 724,328 |
| Length | 200 |
| Max. P | 0.999999 |

| Location | 724,128 – 724,248 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -38.60 |
| Energy contribution | -37.43 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724128 120 + 1 GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUA ((.(((((.((...(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).))))))).((((....)))))).))).)).)).. ( -40.70) >pel_1 81 120 + 1 GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUG ...(((....))).(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).))))))).((((....))))..((((....)))) ( -38.80) >pf5_1 79 120 + 1 GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUG ..(((...((((..(((((((..........((..(....)..)).....((((((((((((........)))))...))))))).))))))).((((....)))))))).)))...... ( -41.20) >pfs_1 81 120 + 1 GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUG ....(((((........)))))((((..(((((..(....)..)......((((((((((((........)))))...)))))))..))))...((((....))))..))))........ ( -38.90) >ppk_1 81 120 + 1 GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCACAAGCUG ...(((..((((..(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).))))))).((((....)))))))))))....... ( -40.70) >pst_1 76 120 + 1 GGAUGCCUCGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUG ....(((((........)))))((((..(((((..(....)..)......(((((((.((((........))))....)))))))..))))...((((....))))..))))........ ( -40.70) >consensus GGAUGCCUUGGCAGUCAGAGGCGAUGAAAGACGUGGUAGCCUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGAUG ...(((....))).(((((((..........((..(....)..)).....(((((((.((((........))))....))))))).))))))).((((....))))..((((....)))) (-38.60 = -37.43 + -1.16)

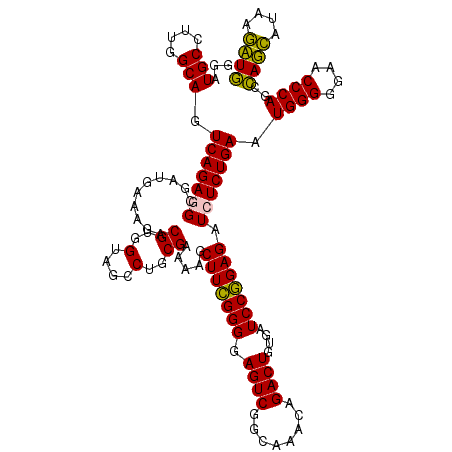
| Location | 724,168 – 724,288 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.33 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -44.51 |
| Energy contribution | -41.18 |
| Covariance contribution | -3.33 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.71 |
| Structure conservation index | 1.07 |
| SVM decision value | 6.04 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724168 120 + 1 CUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCA ..((.....))((((((((.((((((((.....))))..))))...)))))))).(((....)))(((((((....))))))).(((((((((.........)))))))))((....)). ( -43.10) >pel_1 121 120 + 1 CUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCA ..((.....))....((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)). ( -41.40) >pf5_1 119 120 + 1 CUGCGAUAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCA ..((.....))((((((((.((((((((.....))))..))))...)))))))).(((....)))(((((((....))))))).(((((((((.........)))))))))((....)). ( -41.20) >pfs_1 121 120 + 1 CUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUUUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUAAGCUGAAUACAUAGGCUUAAGAGGCGAACCA .(((((....((((((((((((........)))))...)))))))...)).....(((....)))(((((((....))))))).(((((((((.........))))))))).)))..... ( -41.90) >ppk_1 121 120 + 1 CUGCGAUAAGCUUUGGGGAGUCGGCAAACAGACUGUGAUCCAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCACAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCA ..((.....))....((...(((.(...((((....((((....))))))))...(((....)))(((((((....))))))).(((((((((.........)))))))))).))).)). ( -41.10) >pst_1 116 120 + 1 CUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAGAGGCGAACCA .(((((....(((((((.((((........))))....)))))))...)).....(((....)))(((((((....))))))).(((((((((.........))))))))).)))..... ( -41.30) >consensus CUGCGAAAAGCUUCGGGGAGUCGGCAAACAGACUGUGAUCCGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCA .(((((....(((((((.((((........))))....)))))))...)).....(((....)))(((((((....))))))).(((((((((.........))))))))).)))..... (-44.51 = -41.18 + -3.33)

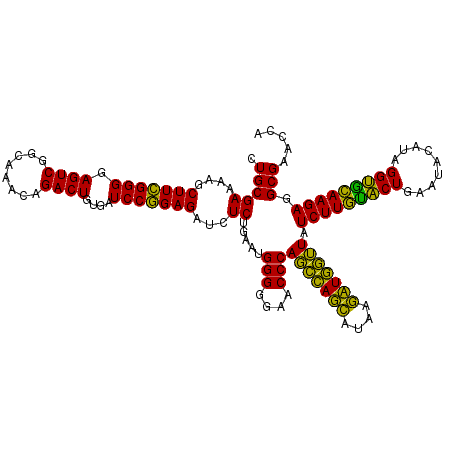
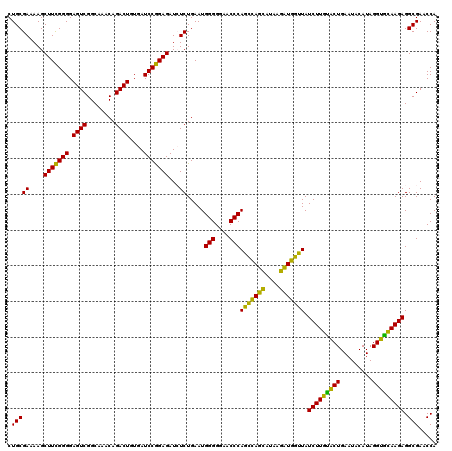
| Location | 724,208 – 724,328 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -41.82 |
| Energy contribution | -38.72 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.51 |
| Structure conservation index | 1.07 |
| SVM decision value | 6.71 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724208 120 + 1 CGGAGAUCUCUGAAUGGGGGAACCCACCUAGGAUAACCUAGGUAUCUUGUACUGAAUCCAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ((((....))))...(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..)).......... ( -39.70) >pel_1 161 120 + 1 CAGAGAUCUCUGAAUGGGGGAACCCACUCAGCAUAAGCUGAGUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ((((....))))...(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..)).......... ( -40.80) >pf5_1 159 120 + 1 CGGAGAUCUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ((((....))))...(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..)).......... ( -37.80) >pfs_1 161 120 + 1 CGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUAAGCUGAAUACAUAGGCUUAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ......((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))..((.((((.(((..........))).))))..)).......... ( -37.50) >ppk_1 161 120 + 1 CAGAGAUCUCUGAAUGGGGGAACCCAGCCAGCACAAGCUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ((((....))))...(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..)).......... ( -40.50) >pst_1 156 120 + 1 CGGAGAUGUCUGAAUGGGGGAACCCAGCCAUCAUAAGAUGGUUAUCUUACACUGAAUACAUAGGUGUAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ......((((.....(((....)))(((((((....))))))).(((((((((.........)))))))))))))..((.((((.(((..........))).))))..)).......... ( -37.60) >consensus CGGAGAUCUCUGAAUGGGGGAACCCAGCCAGCAUAAGAUGGUUAUCUUGUACUGAAUACAUAGGUGCAAGAGGCGAACCAGGGGAACUGAAACAUCUAAGUACCCUGAGGAAAAGAAAUC ((((....))))...(((....)))(((((((....))))))).(((((((((.........)))))))))......((.((((.(((..........))).))))..)).......... (-41.82 = -38.72 + -3.10)

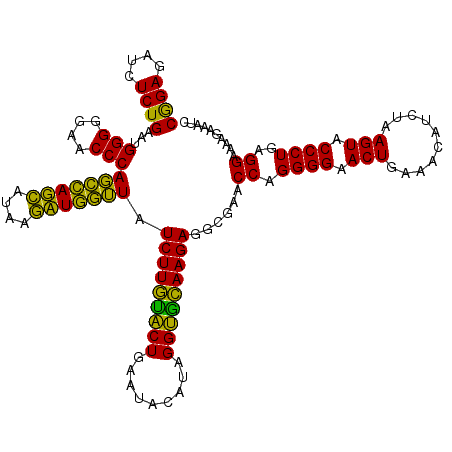
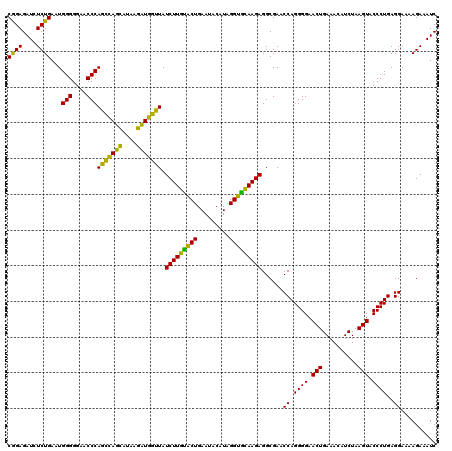
| Location | 724,208 – 724,328 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -29.60 |
| Energy contribution | -27.88 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 724208 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGGAUUCAGUACAAGAUACCUAGGUUAUCCUAGGUGGGUUCCCCCAUUCAGAGAUCUCCG (((((((..(((..((((((...((.((..((((.....))))..)).))..))).)))..)))..........((.(((((((....)))))))(((....)))..))))))))).... ( -33.70) >pel_1 161 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUACUCAGCUUAUGCUGAGUGGGUUCCCCCAUUCAGAGAUCUCUG (((((((.....((((((.(((..((....)))))..))))))...((((((((.((...........)).)))))((((((((....)))))))))))..........))))))).... ( -33.10) >pf5_1 159 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAGAGAUCUCCG (((((((........(((.((((((..(..((((.....))))..)...(((((.((...........)).)))))...(((((....))))))))))).)))......))))))).... ( -28.44) >pfs_1 161 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUAAGCCUAUGUAUUCAGCUUAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAGACAUCUCCG (((.(((........(((.((((((..(..((((.....))))..)...((((((((...........))))))))...(((((....))))))))))).)))......))).))).... ( -29.34) >ppk_1 161 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUGUGCUGGCUGGGUUCCCCCAUUCAGAGAUCUCUG (((((((.....((((((.(((..((....)))))..))))))...(((...((((....))))...((((((((........)))))))))))((((....))))...))))))).... ( -33.50) >pst_1 156 120 - 1 GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUACACCUAUGUAUUCAGUGUAAGAUAACCAUCUUAUGAUGGCUGGGUUCCCCCAUUCAGACAUCUCCG (((.(((........(((.((((((..(..((((.....))))..)...((((((((...........))))))))...(((((....))))))))))).)))......))).))).... ( -29.44) >consensus GAUUUCUUUUCCUCAGGGUACUUAGAUGUUUCAGUUCCCCUGGUUCGCCUCUUGCACCUAUGUAUUCAGUACAAGAUAACCAGCUUAUGAUGGCUGGGUUCCCCCAUUCAGAGAUCUCCG (((((((.....((((((.(((..((....)))))..))))))......(((((.((...........)).)))))...(((((....))))).((((....))))...))))))).... (-29.60 = -27.88 + -1.72)
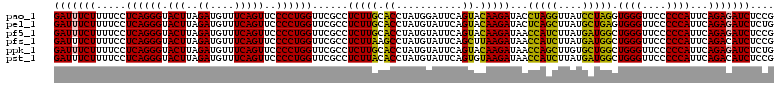
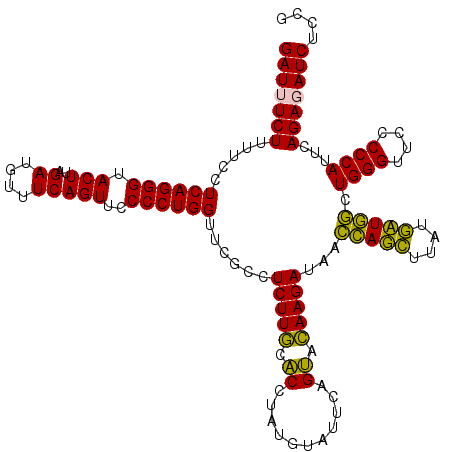
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:46:06 2007