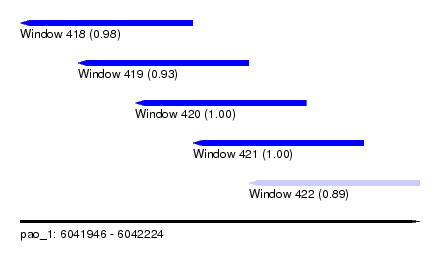
| Sequence ID | pao_1 |
|---|---|
| Location | 6,041,946 – 6,042,224 |
| Length | 278 |
| Max. P | 0.999644 |

| Location | 6,041,946 – 6,042,066 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.44 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -36.17 |
| Energy contribution | -35.78 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041946 120 - 1 CGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAUGACUUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((..((((((((...((((((......))))))....))))...(((.((.((........)).))))).)).))..))))).))).......(((.....)))))).... ( -36.80) >pfs_1 2119 120 - 1 CGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... ( -37.10) >ppk_1 2120 120 - 1 CGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... ( -37.50) >psp_1 2117 120 - 1 CGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... ( -37.10) >pss_1 2118 120 - 1 CGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... ( -37.10) >pst_1 2118 120 - 1 CGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... ( -37.10) >consensus CGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAUGACCUGUGGAUCGGAGUGAAAGGCUAAUCAAGCUCGGAGA (((((((((((((((((((((...((((((......))))))....))))..........(((((((....))))))).)).))))))))).))).......(((.....)))))).... (-36.17 = -35.78 + -0.39)
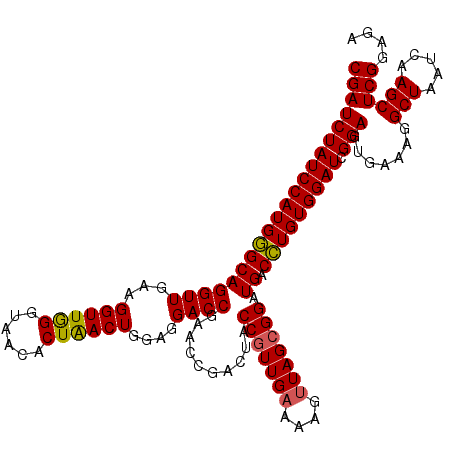
| Location | 6,041,986 – 6,042,105 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.31 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -37.06 |
| Energy contribution | -37.08 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041986 119 - 1 AUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCCACUCCCGUUGAAAAGGUAGGGGAU ....(((.-....)))(((.((.(((((((((....))))...)))))))...))).((((...((((((......))))))....))))...(((.((.((........)).))))).. ( -37.50) >pel_1 2159 120 - 1 AUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ...........((((((((.((.(((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. ( -39.60) >ppk_1 2160 119 - 1 AUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ........-..((((((((.((.(((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. ( -39.60) >psp_1 2157 119 - 1 AUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ........-..((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. ( -39.10) >pss_1 2158 119 - 1 AUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ........-..((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. ( -39.10) >pst_1 2158 119 - 1 AUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ........-..((((((((.(((.((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. ( -39.10) >consensus AUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUAACUGGAGGACCGAACCGACUACCGUUGAAAAGUUAGCGGAU ...........((((((((.((.(((((((((....))))...)))))))...))).((((...((((((......))))))....)))).....)))))(((((((....))))))).. (-37.06 = -37.08 + 0.03)

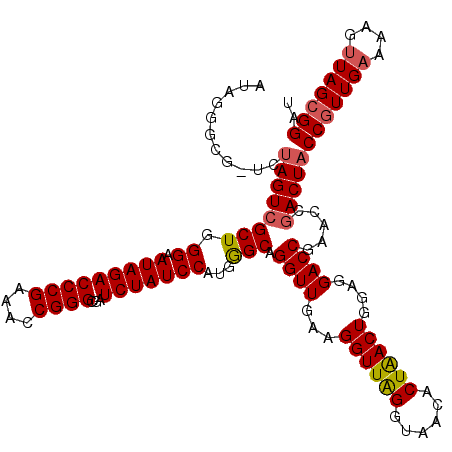
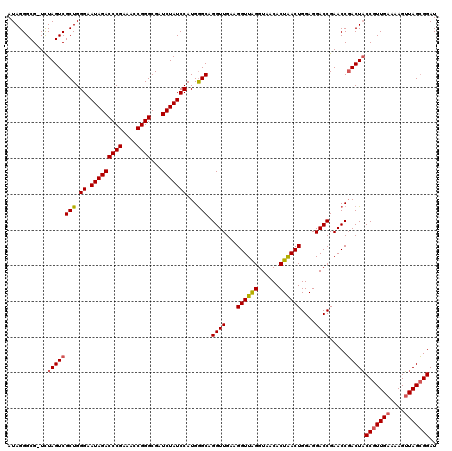
| Location | 6,042,026 – 6,042,145 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.87 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -38.22 |
| Energy contribution | -38.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6042026 119 - 1 GCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGAGCAGGUUGAAGGUUAGGUAACACUGA ((((((((.....((.(((((...((....))..((((.....)))))-)))).))(((.((.(((((((((....))))...)))))))...)))........))))))))........ ( -39.60) >pel_1 2199 120 - 1 GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGA ((((((((.....(((...((.(((((((((((..(((....))).)))))...)))))).)).....))).........((((((..(....)..))))))..))))))))........ ( -40.60) >ppk_1 2200 119 - 1 GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUGA ((((((((.....((.(((((...((....))..))))).....((((-......))))((((....))))....))...((((((..(....)..))))))..))))))))........ ( -40.30) >psp_1 2197 119 - 1 GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAA ((((((((.....((.(((((...((....))..))))).....((.(-((((.((.....)).)))))))....))...((((((..(....)..))))))..))))))))........ ( -39.40) >pss_1 2198 119 - 1 GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAA ((((((((.....((.(((((...((....))..))))).....((.(-((((.((.....)).)))))))....))...((((((..(....)..))))))..))))))))........ ( -39.40) >pst_1 2198 119 - 1 GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUGGGUAACACUAA ((((((((.....((.(((((...((....))..))))).....((.(-((((.((.....)).)))))))....))...((((((..(....)..))))))..))))))))........ ( -39.40) >consensus GCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGGCGAUCUAUCCAUGGGCAGGUUGAAGGUUAGGUAACACUAA ((((((((........(((((...((....))..))))).....(((......)))(((.((.(((((((((....))))...)))))))...)))........))))))))........ (-38.22 = -38.00 + -0.22)
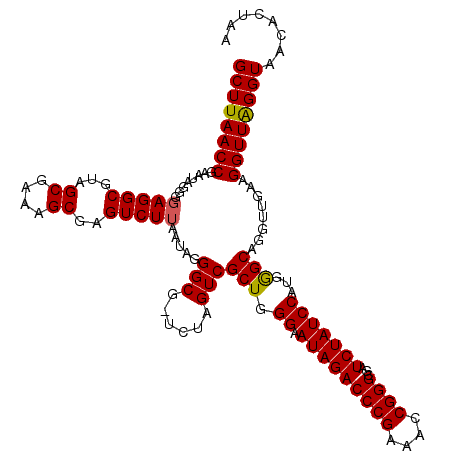
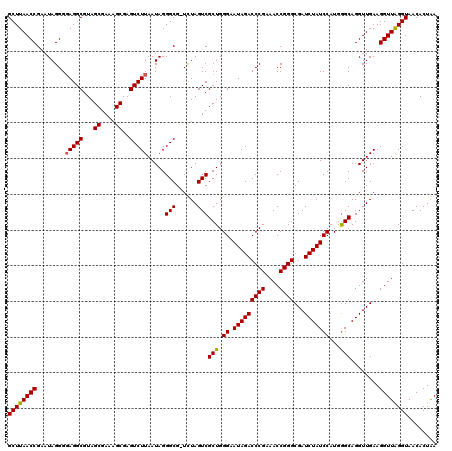
| Location | 6,042,066 – 6,042,185 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.21 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -40.68 |
| Energy contribution | -40.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6042066 119 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGG ............((((((.......(((((((((((((..(((((.((.....)).)))))...((....))..((((.....)))).-....)))))))))))))))))))........ ( -40.21) >pel_1 2239 120 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCGCUUUAGUCGCUGGGUAUAGACCCGAAACCGGG ............((((((.......(((((((((((((..((((..((.....))..))))((.((....))..((((.....))))))....)))))))))))))))))))........ ( -41.21) >pf5_1 2236 119 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGG ............((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))))........ ( -40.91) >ppk_1 2240 119 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UUUAGUCGCUGGGUAUAGACCCGAAACCGGG ............((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....)))).-....)))))))))))))))))))........ ( -40.91) >pss_1 2238 119 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGG ............((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))))........ ( -40.91) >pst_1 2238 119 - 1 CCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG-UCUAGUCGCUGGGAAUAGACCCGAAACCGGG ............((((((.......((((..(((((((..((((..((.....))..))))...((....))((((((.....)))).-))..)))))))..))))))))))........ ( -40.91) >consensus CCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUAAUAGGGCG_UCUAGUCGCUGGGAAUAGACCCGAAACCGGG ............((((((.......(((((((((((((..((((..((.....))..))))...((....))..((((.....))))......)))))))))))))))))))........ (-40.68 = -40.18 + -0.50)

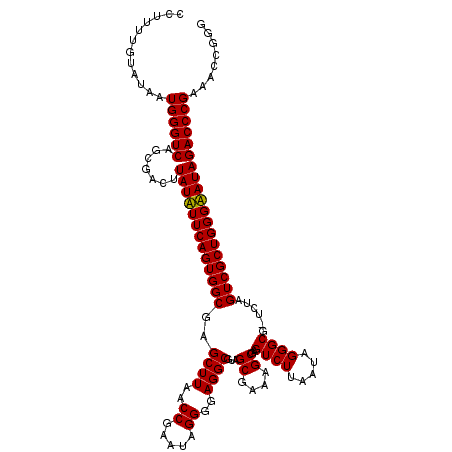
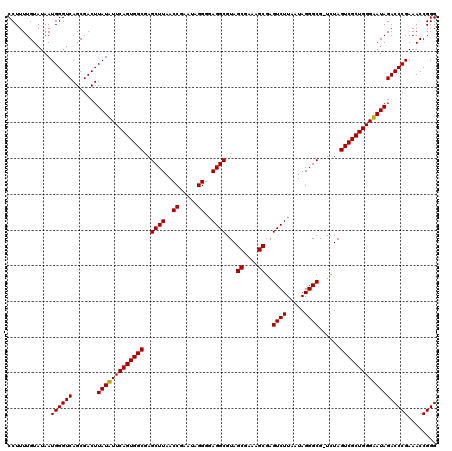
| Location | 6,042,105 – 6,042,224 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -36.27 |
| Energy contribution | -35.22 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.85 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6042105 119 - 1 GUACAAGCAGUGGGAGCCUAC-UUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCAAGCUUAACCGUAUAGGGUAGGCGUAGCGAAAGCGAGUCUUA ...(.((((((((....))))-.)))).)..((((((.(((((..(((((..(((((..((.(((.......))).))..)))))...)))))))))).))...((....)).))))... ( -35.60) >pel_1 2279 120 - 1 GUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA (((((((..(..(..(((((......)))))..)..)......)))))))..(((((..((.(((.......))).))..))))).((.....)).(((((...((....))..))))). ( -35.70) >pf5_1 2275 120 - 1 GUACAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCAAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA ((((..(((((....(((((......))))).)))))))))...(((...((((((((...))))))))..))).(((..((((..((.....))..))))...((....))..)))... ( -35.90) >pfo_1 2277 120 - 1 GUACAAGCAGUGGGAGCAGACUUUGUUCUGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA ......((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))... ( -36.10) >ppk_1 2279 120 - 1 GUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA (((((((..(..(..(((((......)))))..)..)......)))))))..(((((..((.(((.......))).))..))))).((.....)).(((((...((....))..))))). ( -35.70) >pst_1 2277 120 - 1 GUACAAGCAGUGGGAGCCCACUUUGUUGGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUUUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA ......((.(..(..(((((......)))))..)..)(.((((..........))))).))((((..(((((....((..((((..((.....))..))))...)))))))..))))... ( -38.20) >consensus GUACAAGCAGUGGGAGCCUACUUUGUUAGGUGACUGCGUACCUUUUGUAUAAUGGGUCAGCGACUUAUAUUCAGUGGCGAGCUUAACCGAAUAGGGGAGGCGUAGCGAAAGCGAGUCUUA ......((.(..(..(((((......)))))..)..)(.((((..........))))).))(((((..............((((..((.....))..))))...((....)))))))... (-36.27 = -35.22 + -1.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:54:49 2007