
| Sequence ID | pao_1 |
|---|---|
| Location | 6,041,188 – 6,041,826 |
| Length | 638 |
| Max. P | 0.999993 |

| Location | 6,041,188 – 6,041,308 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -20.83 |
| Energy contribution | -19.70 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
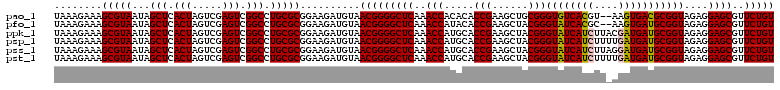
>pao_1 6041188 120 + 1 CAUGGCAUCAACCACUUCGUCAUCUAAAAGACGACUCGUCAUCAGCUCUCGGCCUUGAAACCCCGGAUUUACCUAAGAUUUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCU ..((((..........(((((........)))))...(((..........(((..(((((.(..((.....))...).))))))))......(........)))).......)))).... ( -20.60) >pel_1 1361 119 + 1 CAUGGCAUCAACCACUUCGCGCUCUAAU-GAGCACUCGUCAUCAGCUCUCGGCCUUGAGAUCCCGGAUUUGCCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCU ..((((..............((((....-))))....(((..........(((..(((((((..((.....))...))))))))))......(........)))).......)))).... ( -28.90) >pf5_1 1358 120 + 1 CAUGGCAUCAACCACUUCGUCACCUAAAAGGUAACUCGUCAUCAGCUCUCGGCCUUAGAACCCCGGAUUUACCUAAGAUUCCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCU ..((((............((.(((.....))).))..(((....((.....))...........(((.((......)).)))....................))).......)))).... ( -17.60) >pfs_1 1361 120 + 1 CAUGGCAUCAACCACUUCGUUAACUAAAAGUUAACUCGUCAUCAGCUCUCGGCCUUAAGAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCU ..((((............((((((.....))))))..(((..........(((....(((((..((.....))...)))))..)))......(........)))).......)))).... ( -22.60) >ppk_1 1361 119 + 1 CAUGGUAUCAACCACUUCGUCGCCUAAA-GGCAACUCGUCAUCAGCUCUCGGCCUUGAAAUCCCGGAUUUGCCUAAGAUUUCAGCCUACCACCUUAAACCUGGACAACCAACGCCAGGCU ..(((((...........((.(((....-))).))...............(((..(((((((..((.....))...))))))))))))))).......(((((.(.......)))))).. ( -28.50) >pst_1 1358 120 + 1 CAUGGCAUCAACCACUUCAUGUUCUAAAAGAACACUCGUCAUCAGCUCUCGGCCUUAAGAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACCUGGACUACCAACGCCAGGCU ..(((..............(((((.....)))))................(((....(((((..((.....))...)))))..)))..))).......(((((.(.......)))))).. ( -21.80) >consensus CAUGGCAUCAACCACUUCGUCAUCUAAAAGACAACUCGUCAUCAGCUCUCGGCCUUAAAAUCCCGGAUUUACCUAAGAUCUCAGCCUACCACCUUAAACUUGGACAACCAACGCCAAGCU ..((((............((((((.....))))))..(((..........(((....(((((..((.....))...)))))..)))......(........)))).......)))).... (-20.83 = -19.70 + -1.13)

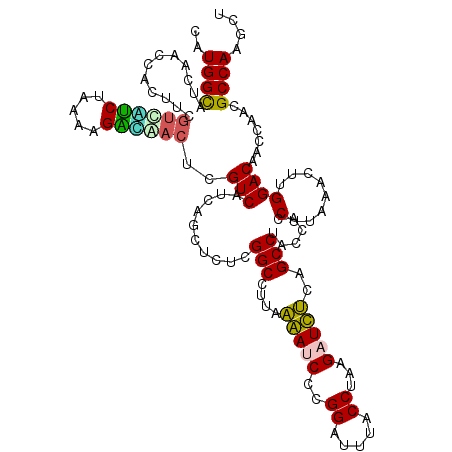

| Location | 6,041,188 – 6,041,308 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -44.67 |
| Energy contribution | -40.82 |
| Covariance contribution | -3.85 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.64 |
| Structure conservation index | 1.07 |
| SVM decision value | 5.74 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041188 120 - 1 AGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAUGACGAGUCGUCUUUUAGAUGACGAAGUGGUUGAUGCCAUG (((((((((.....)).)))))))....((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..)))))). ( -39.90) >pel_1 1361 119 - 1 AGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUGCUC-AUUAGAGCGCGAAGUGGUUGAUGCCAUG (((((((((.....)).)))))))....(((((((((((((((((..((....))..)))))))..)))).......(((.((..((((((-....))))))...)).)))..)))))). ( -44.60) >pf5_1 1358 120 - 1 AGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAUGACGAGUUACCUUUUAGGUGACGAAGUGGUUGAUGCCAUG (((((((((.....)).)))))))....((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..)))))). ( -40.80) >pfs_1 1361 120 - 1 AGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUUAACUUUUAGUUAACGAAGUGGUUGAUGCCAUG (((((((((.....)).)))))))....((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..)))))). ( -39.30) >ppk_1 1361 119 - 1 AGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAUGACGAGUUGCC-UUUAGGCGACGAAGUGGUUGAUACCAUG (((((((((.....)).)))))))....(((((((((((((((((..((....))..)))))))..)))).......(((.((..((((((-....))))))...)).)))..)))))). ( -44.30) >pst_1 1358 120 - 1 AGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAUGACGAGUGUUCUUUUAGAACAUGAAGUGGUUGAUGCCAUG ((((((((.......).)))))))....((((((((((((((((((.((.....))))))))))..)))).......(((.((..((((((.....))))))...)).)))..)))))). ( -41.90) >consensus AGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUCUCAAGGCCGAGAGCUGAUGACGAGUUAUCUUUUAGAUGACGAAGUGGUUGAUGCCAUG (((((((((.....)).)))))))....(((((((((((((((((..((....))..)))))))..)))).......(((.((..((((((.....))))))...)).)))..)))))). (-44.67 = -40.82 + -3.85)
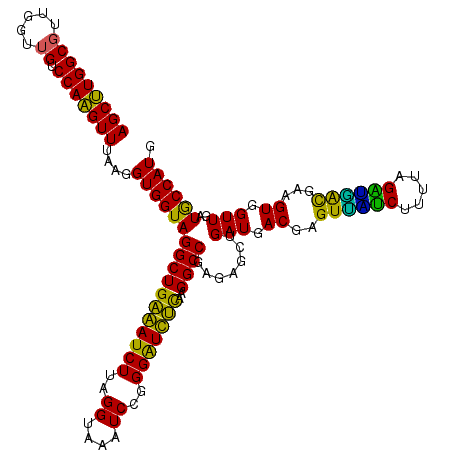
| Location | 6,041,228 – 6,041,348 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -39.75 |
| Consensus MFE | -41.01 |
| Energy contribution | -39.02 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041228 120 - 1 CUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAU .((((...(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))((((((((((((.((.....))))))))))..)))).))))..... ( -39.00) >pel_1 1400 120 - 1 CUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAGAUCUUAGGCAAAUCCGGGAUCUCAAGGCCGAGAGCUGAU ....(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....)))).. ( -42.80) >pf5_1 1398 120 - 1 CUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGGAAUCUUAGGUAAAUCCGGGGUUCUAAGGCCGAGAGCUGAU ((((..((....))...))))((((((..((.((((((....)))))).))..))))))........(((....((((((((((((.((.....))))))))))..))))....)))... ( -38.90) >pfo_1 1400 120 - 1 CUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGGUUUCAAGGCCGAGAGCUGAU (((((.((....)).).))))((((((..((.((((((....)))))).))..))))))........(((....((((((((((((.((.....))))))))))..))))....)))... ( -38.00) >ppk_1 1400 120 - 1 CUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGAAAUCUUAGGCAAAUCCGGGAUUUCAAGGCCGAGAGCUGAU ....(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....)))).. ( -40.30) >pst_1 1398 120 - 1 CUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGAGAUCUUAGGUAAAUCCGGGAUCUUAAGGCCGAGAGCUGAU (((((.((....)).).))))((((....((.((((((....)))))).))....))))........(((....((((((((((((.((.....))))))))))..))))....)))... ( -39.50) >consensus CUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGAAAUCUUAGGUAAAUCCGGGAUUUCAAGGCCGAGAGCUGAU ....(((((((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))(((((((((((..((....))..)))))))..))))....)))).. (-41.01 = -39.02 + -1.99)

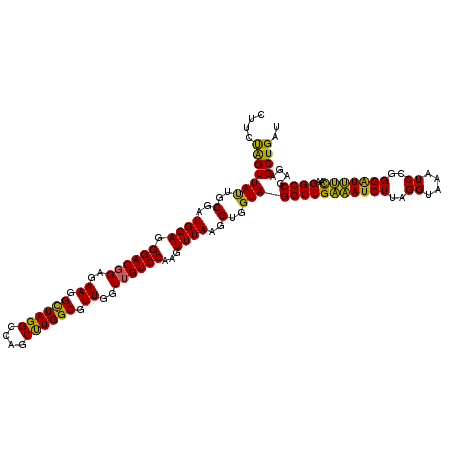
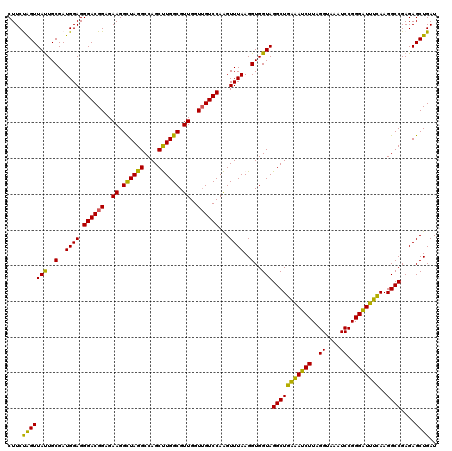
| Location | 6,041,268 – 6,041,388 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -38.35 |
| Energy contribution | -37.13 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041268 120 - 1 GAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGA ......((((((.(((.(((((((((........)))))..)))).)))))))))......((((((..((.((((((....)))))).))..)))))).((((((......)))))).. ( -39.90) >pel_1 1440 120 - 1 GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGA ........(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))). ( -38.70) >pf5_1 1438 120 - 1 GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCUUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGG ........(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))). ( -36.40) >ppk_1 1440 120 - 1 GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGA ........(((((..(((((.(((((........)))))..)))))..(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))). ( -38.90) >pss_1 1438 120 - 1 GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAGGUUUAAGGUGGUAGGCUGA ........(((((...((((.(((((........)))))..))))...(((..(..((((.((((((..((.((((((....)))))).))..))))))...))))..)..)))))))). ( -37.90) >pst_1 1438 120 - 1 GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUAGUCCAGGUUUAAGGUGGUAGGCUGA ........(((((...((((.(((((........)))))..))))...(((..(..((((.((((....((.((((((....)))))).))....))))...))))..)..)))))))). ( -35.00) >consensus GAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCCAGCCUGGCGUUGGUUGUCCAAGUUUAAGGUGGUAGGCUGA ......((((((...(((((.(((((........)))))..)))))...))))))......((((((..((.((((((....)))))).))..)))))).((((((......)))))).. (-38.35 = -37.13 + -1.22)

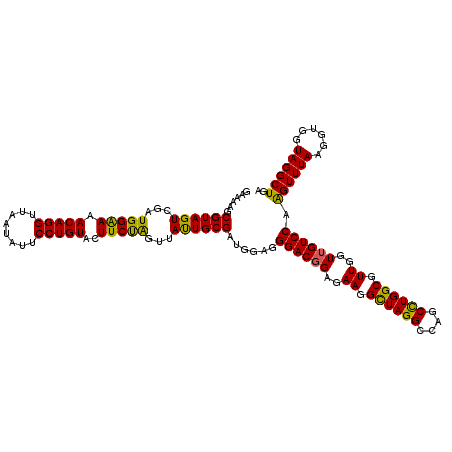
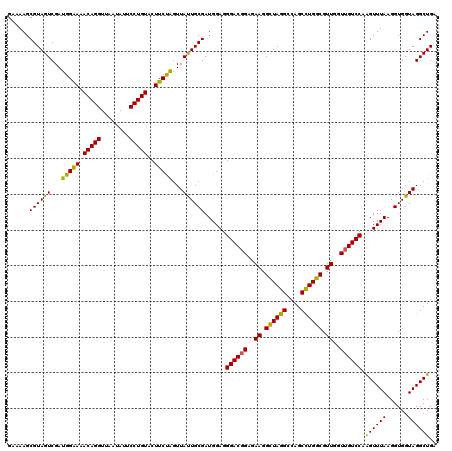
| Location | 6,041,308 – 6,041,428 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -36.72 |
| Energy contribution | -35.87 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041308 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUACUUCUGGUUACUGCGAUGGAGGGACGGAGAAGGCUAGGCC ..........((..((((..((.((((((....(...(((....)))(((((.(((.(((((((((........)))))..)))).)))))))))....)))))).))...))))..)). ( -38.50) >pel_1 1480 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCC ..........((..((((..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...))))..)). ( -39.90) >pf5_1 1478 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCC ..........((..((((..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...))))..)). ( -37.60) >ppk_1 1480 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCCAGUUAUUGCGAUGGAGGGACGGAGAAGGUUAGGCC .(((((((((.....)))..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...))))))... ( -38.00) >psp_1 1477 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCC (((.((((((...(.(((..((((((.(......).))))))..))).).))))))....))).((((....(((((((.(((((.((....)).).))))..)))).))).....)))) ( -32.80) >pst_1 1478 120 - 1 GUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUGGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCC ..........((..((((..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...))))..)). ( -36.30) >consensus GUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUACUUCUAGUUAUUGCGAUGGAGGGACGGAGAAGGCUAGGCC ..........((..((((..((.((((((....(...(((....)))(((((...(((((.(((((........)))))..)))))...))))))....)))))).))...))))..)). (-36.72 = -35.87 + -0.86)

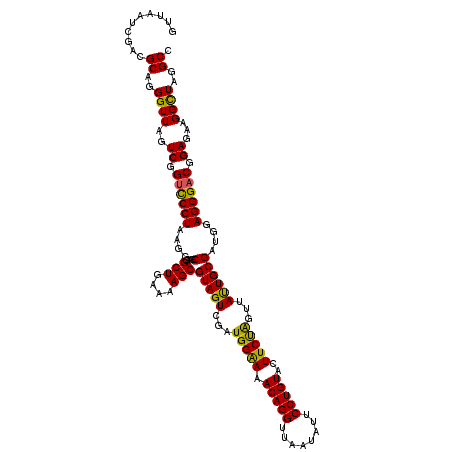
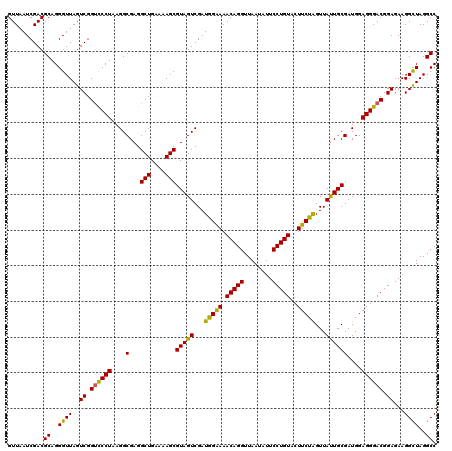
| Location | 6,041,348 – 6,041,468 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -36.35 |
| Energy contribution | -37.05 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041348 120 + 1 UACAGGAAUAUUAACCUGUUUCCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUU .(((((........)))))....................((.((.((((..((((.((((.....((((((.((((....)))))))))).....))))))))..)))).)).))..... ( -38.10) >pel_1 1520 120 + 1 UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUU .(((((........)))))........((((.(((..........((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)))))))... ( -38.90) >ppk_1 1520 120 + 1 UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUU .(((((........)))))........((((.(((..........((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)))))))... ( -38.90) >psp_1 1517 120 + 1 UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUU .(((((........)))))..........(((((((.....(((((......))))).((((((.((((((.((((....)))))))))).....))))))....)))))))........ ( -40.30) >pss_1 1518 120 + 1 UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUU .(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).))..... ( -40.60) >pst_1 1518 120 + 1 UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUU .(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).))..... ( -40.60) >consensus UACAGGAAUAUUAACCUGUUUUCCAUCGACUACGCUUUUCAGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUU .(((((........)))))....................((.((.((((..(((((((((.....((((((.((((....)))))))))).....))))))))).)))).)).))..... (-36.35 = -37.05 + 0.70)
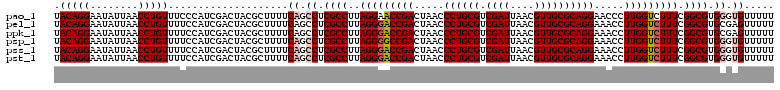
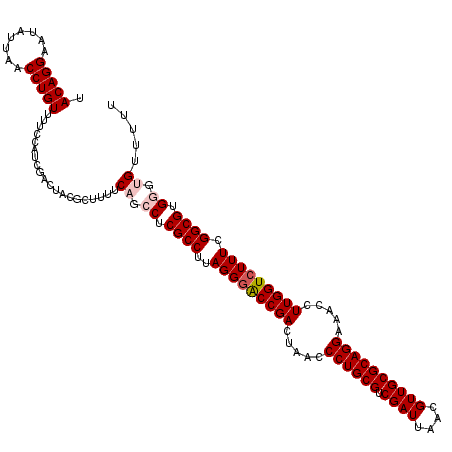
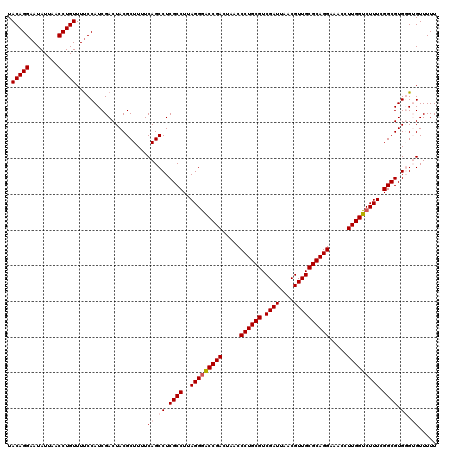
| Location | 6,041,348 – 6,041,468 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -35.04 |
| Energy contribution | -35.07 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041348 120 - 1 AAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGGAAACAGGUUAAUAUUCCUGUA .......((((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........))))..(((((........))))). ( -39.30) >pel_1 1520 120 - 1 AAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ......((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).)))......(((((........))))). ( -37.00) >ppk_1 1520 120 - 1 AAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ......((((.((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))...).)))......(((((........))))). ( -37.00) >psp_1 1517 120 - 1 AAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ............(((((.((((....))))((((((...((.....)).)))))).....))))).((...(((.(.(((....))).).)))...))...(((((........))))). ( -37.40) >pss_1 1518 120 - 1 AAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ........(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........))))). ( -37.60) >pst_1 1518 120 - 1 AAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ........(((((((....((((......(((((((...((.....)).)))))))......)))).....))))..(((....)))........)))...(((((........))))). ( -37.60) >consensus AAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCUGAAAAGCGUAGUCGAUGGAAAACAGGUUAAUAUUCCUGUA ............(((((.((((....))))((((((...((.....)).)))))).....))))).((...(((.(.(((....))).).)))...))...(((((........))))). (-35.04 = -35.07 + 0.03)

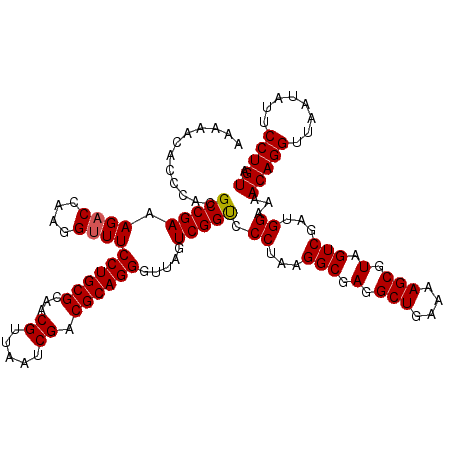
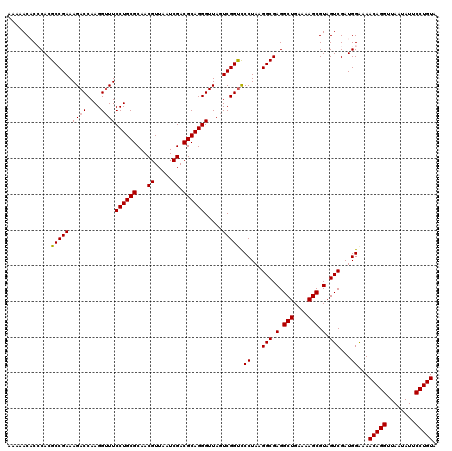
| Location | 6,041,388 – 6,041,508 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -41.31 |
| Energy contribution | -40.45 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.49 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.26 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041388 120 + 1 AGCCUCGCCUUAGGAACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..((((.((((.....((((((.((((....)))))))))).....))))))))..))))(((((((.....))))))).....(((((....)).)))....))...... ( -39.00) >pel_1 1560 120 + 1 AGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))...... ( -40.90) >ppk_1 1560 120 + 1 AGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))...... ( -40.90) >psp_1 1557 120 + 1 AGCCUCGCCUUAGGGGCCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .(((((......))))).((((((.((((((.((((....)))))))))).....))))))...((((((((((((.....))))))).)))))............((....))...... ( -42.00) >pss_1 1558 120 + 1 AGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))...... ( -41.50) >pst_1 1558 120 + 1 AGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))...... ( -41.50) >consensus AGCCUCGCCUUAGGGACCGACUAACCCUGCGUCGAUUAACGUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCU .((..((((..(((((((((.....((((((.((((....)))))))))).....))))))))).))))(((((((.....))))))).....(((((....)).)))....))...... (-41.31 = -40.45 + -0.86)
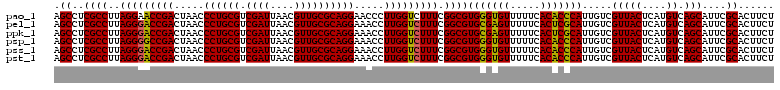
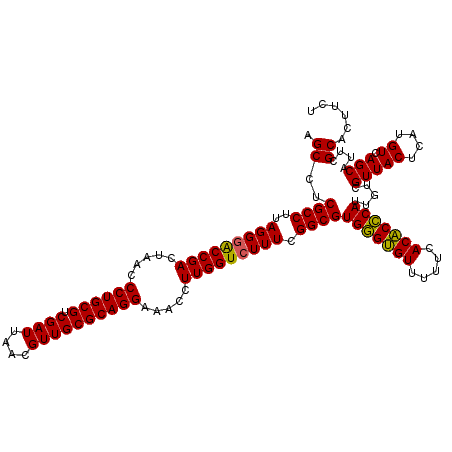
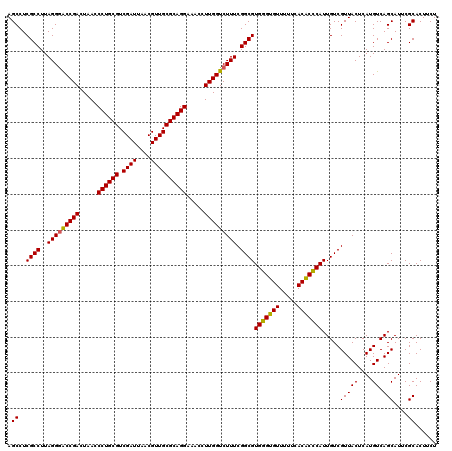
| Location | 6,041,388 – 6,041,508 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -38.19 |
| Energy contribution | -37.33 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.20 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041388 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUUCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... ( -36.80) >pel_1 1560 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... ( -38.00) >ppk_1 1560 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... ( -38.00) >psp_1 1557 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGCCCCUAAGGCGAGGCU .........(((.(((...(((.(((((....(((((((.....))))))).((((..((((....))))..)).))..))))).)))...))).)))((((.(((.....)))..)))) ( -36.30) >pss_1 1558 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... ( -38.60) >pst_1 1558 120 - 1 AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... ( -38.60) >consensus AGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAACGUUAAUCGACGCAGGGUUAGUCGGUCCCUAAGGCGAGGCU ....((.((..((((......)))).))))..(((((((.....)))))))((((....((((......(((((((...((.....)).)))))))......)))).....))))..... (-38.19 = -37.33 + -0.86)
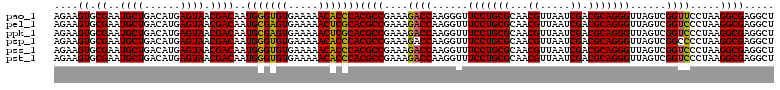
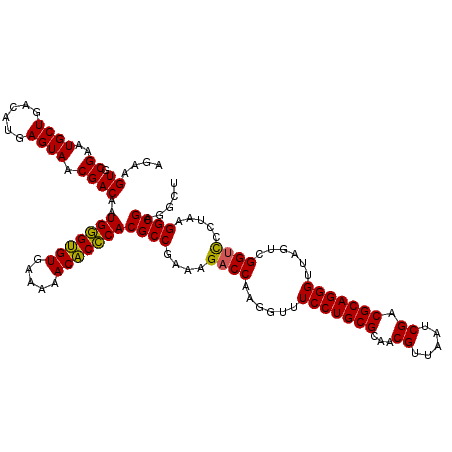
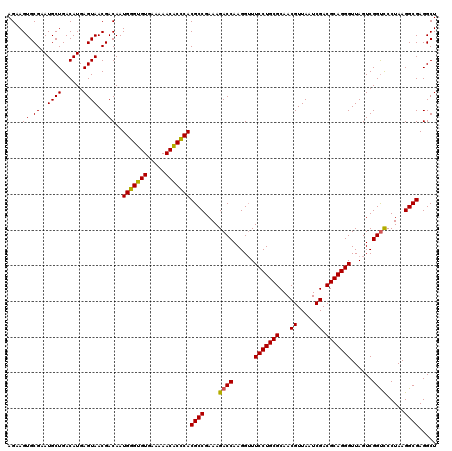
| Location | 6,041,428 – 6,041,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -29.68 |
| Energy contribution | -28.79 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.72 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041428 120 + 1 GUUGCGCAGGAACCCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.99) >pel_1 1600 120 + 1 GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.39) >ppk_1 1600 120 + 1 GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGCGAGUUUUUCACUCGCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.39) >psp_1 1597 120 + 1 GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.99) >pss_1 1598 120 + 1 GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.99) >pst_1 1598 120 + 1 GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) ( -28.99) >consensus GUUGCGCAGGAAACCUUGGUCUUUCGGCGUGGGUGUUUUUCACACCCAUUGUCGUUACUCAUGUCAGCAUUCGCACUUCUGAUACCUCCAGCAAGCUUCUCAACUCACCUUCACAGGCUU ((((.(.(((.......(((....((((((((((((.....))))))).)))))..)))..((((((.(........)))))))))))))))((((((................)))))) (-29.68 = -28.79 + -0.89)

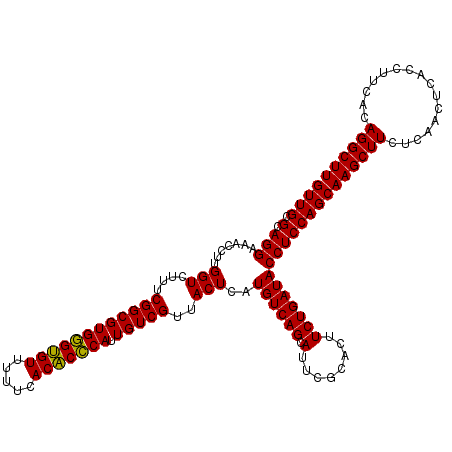
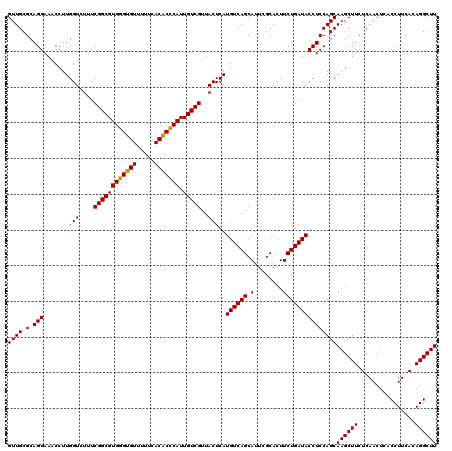
| Location | 6,041,428 – 6,041,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -33.20 |
| Energy contribution | -32.48 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.68 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041428 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGGUUCCUGCGCAAC ..((((.(...((..((((....))))..))).)))).......((((((((.((.................(((((((.....)))))))...(....)....)).))))..))))... ( -34.00) >pel_1 1600 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... ( -32.60) >ppk_1 1600 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUGAAAAACUCGCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... ( -32.60) >psp_1 1597 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... ( -33.20) >pss_1 1598 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... ( -33.20) >pst_1 1598 120 - 1 AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... ( -33.20) >consensus AAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUGAAAAACACCCACGCCGAAAGACCAAGGUUUCCUGCGCAAC ((((((.....((..((((....))))..))...(((.........(((..((((......)))).......(((((((.....))))))))))(....)))).)))))).......... (-33.20 = -32.48 + -0.72)

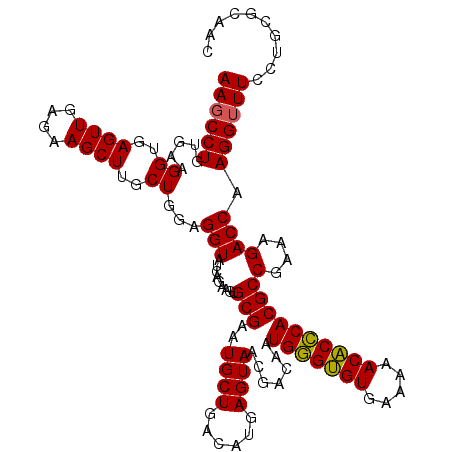
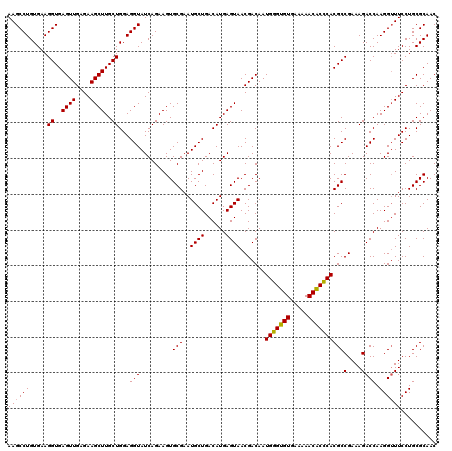
| Location | 6,041,468 – 6,041,586 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.60 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -33.59 |
| Energy contribution | -33.15 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041468 118 - 1 GGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUG ..(((((((..--..)))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))).((((..((.......))..)))). ( -37.30) >pel_1 1640 119 - 1 GGGUAUCACCUU-UGGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUG ..((((((((..-.))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..(((..(((.......)))..))) ( -35.50) >pfo_1 1640 118 - 1 GGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUG ..(((((((..--..)))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))).((((..((.......))..)))). ( -36.80) >ppk_1 1640 120 - 1 GGGUAUCAUCUUACGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGCGAGUG ..((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..(((..(((.......)))..))) ( -35.00) >pss_1 1638 120 - 1 GGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUG ..((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))).((((..((.......))..)))). ( -34.70) >pst_1 1638 120 - 1 GGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUG ..((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...)))).((((..((.......))..)))). ( -34.90) >consensus GGGUAUCACCUU_AGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUCAGAAGUGCGAAUGCUGACAUGAGUAACGACAAUGGGUGUG ..((((((((....))))))))((((.....((((((((...((((.(...((..((((....))))..))).))))..))))).)))...))))..(((..((....)).)))...... (-33.59 = -33.15 + -0.44)

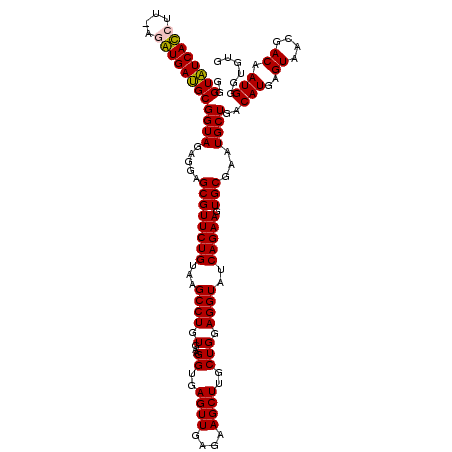
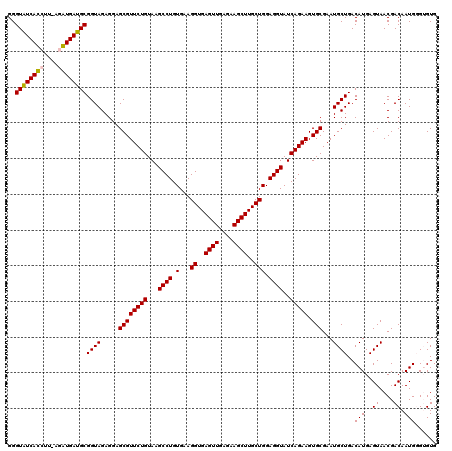
| Location | 6,041,508 – 6,041,626 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -38.24 |
| Energy contribution | -37.85 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041508 118 - 1 CGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -41.40) >pfo_1 1680 118 - 1 CGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -40.70) >ppk_1 1680 120 - 1 CGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUACGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -38.20) >psp_1 1677 120 - 1 CGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -38.80) >pss_1 1678 120 - 1 CGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -38.60) >pst_1 1678 120 - 1 CGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... ( -38.80) >consensus CGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUU_AGAUGAUGCGGUAGAGGAGCGUUCUGUAAGCCUGUGAAGGUGAGUUGAGAAGCUUGCUGGAGGUAUC (....).....(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..)))))..((((.(...((..((((....))))..))).))))... (-38.24 = -37.85 + -0.39)

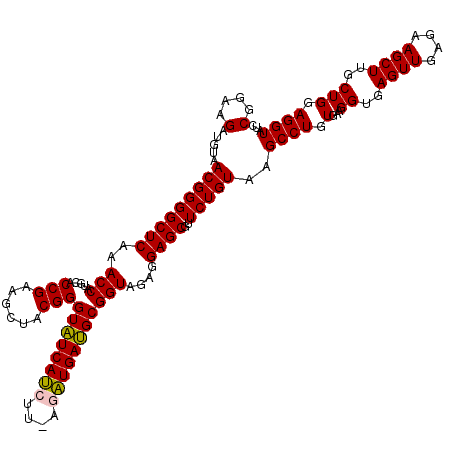
| Location | 6,041,548 – 6,041,666 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.55 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -36.14 |
| Energy contribution | -35.75 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041548 118 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCACACACCGAAGCUGCGGGUGUCACGU--AAGUGACGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..))))) ( -39.30) >pfo_1 1720 118 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUACACCGAAGCUACGGGUAUCACGC--AAGUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))(((((((..--..))))))))))....))))..))))) ( -38.60) >ppk_1 1720 120 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUACGAUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..))))) ( -36.10) >psp_1 1717 120 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..))))) ( -36.70) >pss_1 1718 120 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUAGGAUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..))))) ( -36.50) >pst_1 1718 120 - 1 UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUUUUGAUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..))))) ( -36.70) >consensus UAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCGCGGAAGAUGUAACGGGGCUCAAACCAUGCACCGAAGCUACGGGUAUCAUCUU_AGAUGAUGCGGUAGAGGAGCGUUCUGU ........(((((...(((.(((.....))).))).)))))..........(((((((((..(((.....(((......)))((((((((....)))))))))))....))))..))))) (-36.14 = -35.75 + -0.39)

| Location | 6,041,626 – 6,041,746 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.16 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041626 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGAC ...((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...))))(((((....(((........)))........))))). ( -27.70) >pf5_1 1796 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGAC .(.((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...)))).)..........(((((..............))))). ( -28.24) >pfo_1 1798 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGAC .(.((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...)))).)..........(((((..............))))). ( -28.24) >pfs_1 1799 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGAC .(.((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...)))).)..........(((((..............))))). ( -28.24) >pss_1 1798 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGAC ...((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...))))((((((((.(((........))).)))....))))). ( -31.10) >pst_1 1798 120 + 1 CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGAC ...((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...))))((((((((.(((........))).)))....))))). ( -31.10) >consensus CGCAGGCCGACUCGACUAGUGAGCUAUUACGCUUUCUUUAAAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGAC ...((((.(((..(.((((.((((......)))).......(((.(((((.......))))).))).))))).)))...))))(((((....(((..............)))..))))). (-28.38 = -28.16 + -0.22)

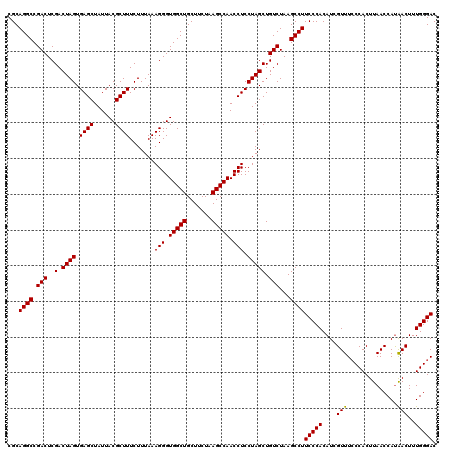
| Location | 6,041,626 – 6,041,746 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -37.49 |
| Consensus MFE | -36.23 |
| Energy contribution | -35.87 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041626 120 - 1 GUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((..(((((.............))))).)))))(((((..(((.(((((((((.(((((.......))))).)))).......(((......)))..)))))...))))))))... ( -36.72) >pf5_1 1796 120 - 1 GUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((...(((....))).)))))......(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..). ( -38.00) >pfo_1 1798 120 - 1 GUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((...(((....))).)))))......(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..). ( -38.00) >pfs_1 1799 120 - 1 GUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((...(((....))).)))))......(..((..(((((.(((.....(((((.(((((.......))))).)))))..((..(((......)))..)).))))))))..))..). ( -38.00) >pss_1 1798 120 - 1 GUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((....(((.(((........))).))))))))(((((..(((.(((((((((.(((((.......))))).)))).......(((......)))..)))))...))))))))... ( -37.10) >pst_1 1798 120 - 1 GUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((....(((.(((........))).))))))))(((((..(((.(((((((((.(((((.......))))).)))).......(((......)))..)))))...))))))))... ( -37.10) >consensus GUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUUUAAAGAAAGCGUAAUAGCUCACUAGUCGAGUCGGCCUGCG .(((((....(((.(((........))).))))))))(((((..(((.(((((((((.(((((.......))))).)))).......(((......)))..)))))...))))))))... (-36.23 = -35.87 + -0.36)

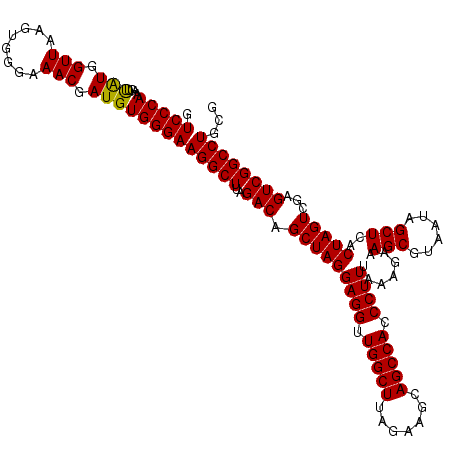
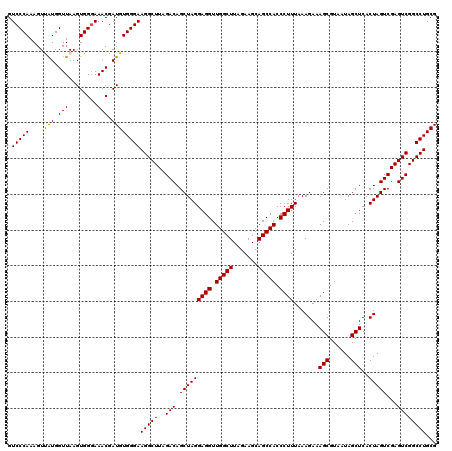
| Location | 6,041,666 – 6,041,786 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -32.82 |
| Energy contribution | -32.46 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041666 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUACCACUUAACCACAACUUUGGGACCUUAGCUGGCGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..(((((....(((........)))........))))).....)))))))).)))))..))).)))))........... ( -31.50) >pf5_1 1836 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).)))))........... ( -32.54) >pfo_1 1838 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).)))))........... ( -32.54) >pfs_1 1839 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).)))))........... ( -32.54) >pss_1 1838 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).)))))........... ( -35.50) >pst_1 1838 120 + 1 AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUGACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..((((((((.(((........))).)))....))))).....)))))))).)))))..))).)))))........... ( -35.50) >consensus AAGGGUGGCUGCUUCUAAGCCAACCUCCUAGCUGUCUAAGCCUUCCCACAUCGUUUCCCACUUAACCAUAACUUUGGGACCUUAGCUGACGGUCUGGGUUGUUUCCCUUUUCACGACGGA (((((.((((.......))))(((..((((((((((..(((..(((((....(((..............)))..))))).....)))))))).)))))..))).)))))........... (-32.82 = -32.46 + -0.36)
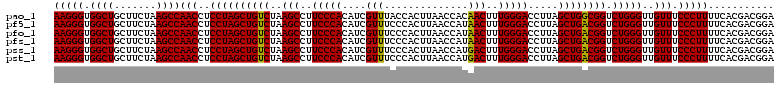
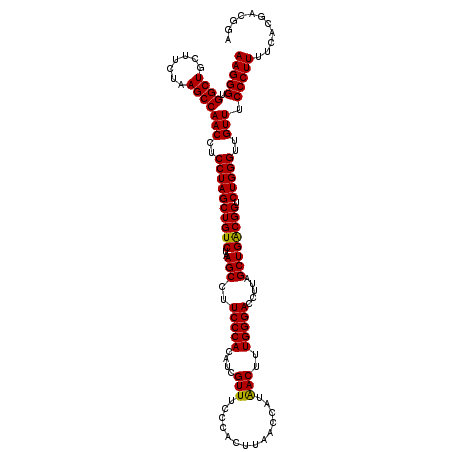
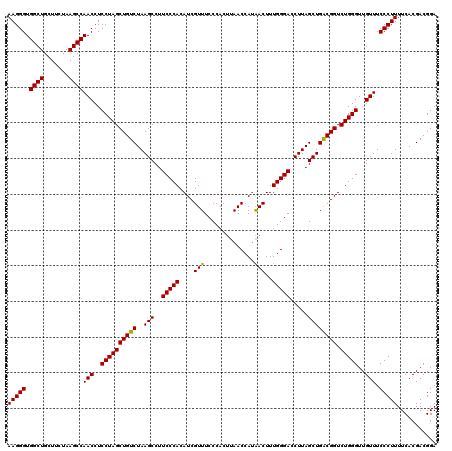
| Location | 6,041,666 – 6,041,786 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -35.86 |
| Energy contribution | -36.08 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041666 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((.....(((......))).((((((.((((..(....)..)))))))))).........)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -36.30) >pf5_1 1836 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -37.30) >pfo_1 1838 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -37.30) >pfs_1 1839 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -37.30) >pss_1 1838 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((.........))))(((......))).....))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -35.70) >pst_1 1838 120 - 1 UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((.........))))(((......))).....))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. ( -35.70) >consensus UCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGGCUUAGACAGCUAGGAGGUUGGCUUAGAAGCAGCCACCCUU ..((((((......(....)..((((((((((.((((..(....)..))))))))))...))))..)))))).(((..((((....(((((....)))))(((....))))))).))).. (-35.86 = -36.08 + 0.22)

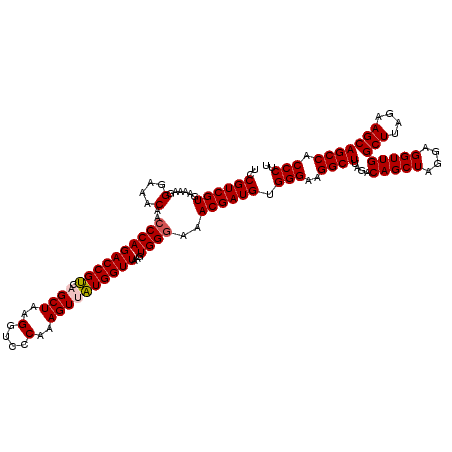
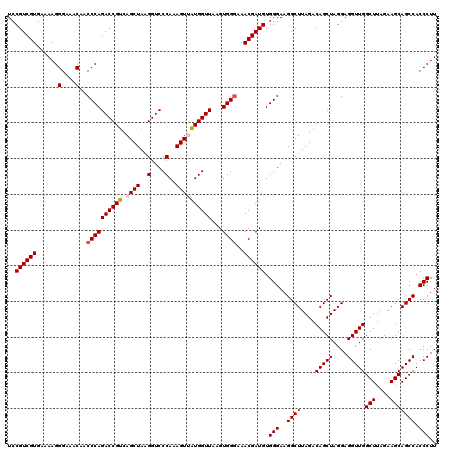
| Location | 6,041,706 – 6,041,826 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -36.64 |
| Energy contribution | -36.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 6041706 120 - 1 AGAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUUGUGGUUAAGUGGUAAACGAUGUGGGAAGG .....(((((....((((.....((((((((..(....)..)))))))).....(....)..))))((((((.((((..(....)..))))))))))...)))))............... ( -38.30) >pf5_1 1876 120 - 1 ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGG ....((((...)))).(....).((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))..((.(....).)).)))).... ( -37.60) >pfo_1 1878 120 - 1 ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGG ....((((...)))).(....).((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))..((.(....).)).)))).... ( -37.60) >pfs_1 1879 120 - 1 ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGG ....((((...)))).(....).((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))..((.(....).)).)))).... ( -37.60) >pss_1 1878 120 - 1 ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGG ...(((..((.(..((((.....((((((((..(....)..)))))))).....(....)..))))..)))...)))....(((((....(((.(((........))).))))))))... ( -37.30) >pst_1 1878 120 - 1 ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGG ...(((..((.(..((((.....((((((((..(....)..)))))))).....(....)..))))..)))...)))....(((((....(((.(((........))).))))))))... ( -37.30) >consensus ACAAGUGCCGAGCAUGGGAGACACACGGCGGGUGCUAACGUCCGUCGUGAAAAGGGAAACAACCCAGACCGUCAGCUAAGGUCCCAAAGUUAUGGUUAAGUGGGAAACGAUGUGGGAAGG ....((((...)))).(....).((((((((..(....)..)))))))).....(....)..((((((((((.((((..(....)..))))))))))..((.(....).)).)))).... (-36.64 = -36.70 + 0.06)
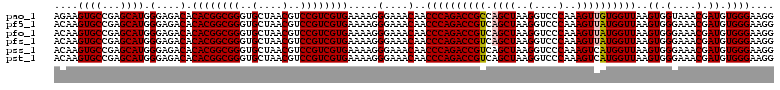
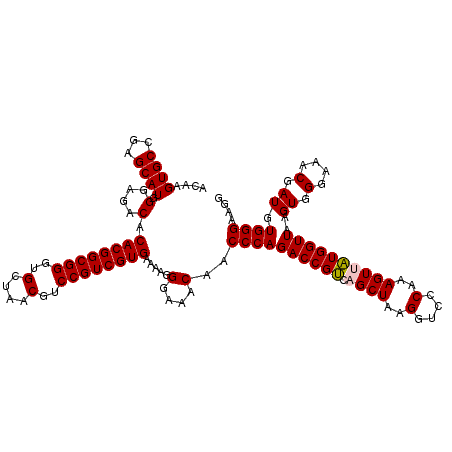
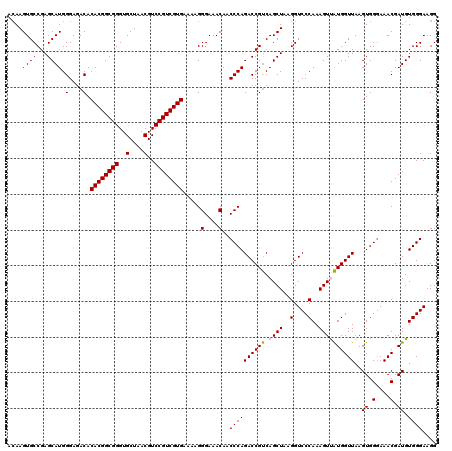
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:54:42 2007