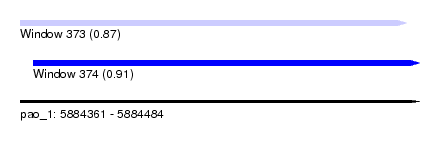
| Sequence ID | pao_1 |
|---|---|
| Location | 5,884,361 – 5,884,484 |
| Length | 123 |
| Max. P | 0.911163 |

| Location | 5,884,361 – 5,884,480 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.39 |
| Mean single sequence MFE | -45.87 |
| Consensus MFE | -37.32 |
| Energy contribution | -36.72 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5884361 119 + 1 GAUAACUGCAACAACGGAGGUUGCCAGUUGGA-CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAGGUCUACUGCAACCGCCACCUUGAACUUUCGGGUUCAAGGGCUAACCCGACAG .......((......((.((((((.(((.(((-(((.(((((......))))).)((....))....)))))))))))))).)).(((((((((....)))))))))))........... ( -45.50) >pst_1 41 120 + 1 GAUACGCACAACCACGGGGGUUGCACGUUGGGGUUGGUGUGCAUGUCCGCUCGACGGAAAGCCUUAAAACCUCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUACACCGACAG .....((........((((((((((....((((((...(.((...((((.....))))..)))....)))))).)))))))))).(((((((((....)))))))))))........... ( -44.10) >pau_1 41 119 + 1 GAUAACUGCAACAACGGAGGUUGCCAGUUGGA-CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAGGUCUACUGCAACCGCCACCUUGAACUUUCGGGUUCAAGGGCUAACCCGACAG .......((......((.((((((.(((.(((-(((.(((((......))))).)((....))....)))))))))))))).)).(((((((((....)))))))))))........... ( -45.50) >pel_1 25 111 + 1 GAUACGCACAACCACGGGGGUUGCACGCUGGGGCCGGUGUGCAUGUCCGCCCGACGGAAAGCCUUAACGCCCCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUA--------- .....((........((((((((((....((((((((.(((......))))))..((....)).....))))).)))))))))).(((((((((....)))))))))))..--------- ( -47.20) >pfs_1 41 119 + 1 GAUUCGCACUACCCUGGAAGUUGCACGUUGGG-CUGGUGUGCAUGUCCGCUAGACGGAAAGCCUUAAAGCCUACUGCAUCUUCCACCUUGAACUUUCGGGUUCAAGGGCUAAGUCGACAG ...(((.(((....((((((.((((.((.(((-((...(.((...((((.....))))..)))....))))))))))).))))))(((((((((....)))))))))....))))))... ( -45.70) >ppk_1 25 111 + 1 GAUACGCACAACCACGGGGGUUGCACGCUGGGGCCGGUGUGCAUGUCCGCCCGACGGAAAGCCUUAACGCCCCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUA--------- .....((........((((((((((....((((((((.(((......))))))..((....)).....))))).)))))))))).(((((((((....)))))))))))..--------- ( -47.20) >consensus GAUACGCACAACCACGGAGGUUGCACGUUGGG_CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAAGCCUACUGCAACCUCCACCUUGAACUUUCGGGUUCAAGGGCUAA_CCGACAG ...............((((((((((....(((.((...(.((...((((.....))))..)))....)))))..)))))))))).(((((((((....)))))))))............. (-37.32 = -36.72 + -0.61)


| Location | 5,884,365 – 5,884,484 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -37.32 |
| Energy contribution | -36.72 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 5884365 119 + 1 ACUGCAACAACGGAGGUUGCCAGUUGGA-CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAGGUCUACUGCAACCGCCACCUUGAACUUUCGGGUUCAAGGGCUAACCCGACAGCGGC .((((......((.((((((.(((.(((-(((.(((((......))))).)((....))....)))))))))))))).)).(((((((((....)))))))))............)))). ( -48.90) >pst_1 45 119 + 1 CGCACAACCACGGGGGUUGCACGUUGGGGUUGGUGUGCAUGUCCGCUCGACGGAAAGCCUUAAAACCUCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUACACCGACAGCGG- (((........((((((((((....((((((...(.((...((((.....))))..)))....)))))).)))))))))).(((((((((....)))))))))............))).- ( -46.50) >pau_1 45 119 + 1 ACUGCAACAACGGAGGUUGCCAGUUGGA-CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAGGUCUACUGCAACCGCCACCUUGAACUUUCGGGUUCAAGGGCUAACCCGACAGCGGC .((((......((.((((((.(((.(((-(((.(((((......))))).)((....))....)))))))))))))).)).(((((((((....)))))))))............)))). ( -48.90) >pel_1 29 107 + 1 CGCACAACCACGGGGGUUGCACGCUGGGGCCGGUGUGCAUGUCCGCCCGACGGAAAGCCUUAACGCCCCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUA------------- .((........((((((((((....((((((((.(((......))))))..((....)).....))))).)))))))))).(((((((((....)))))))))))..------------- ( -47.20) >pfs_1 45 119 + 1 CGCACUACCCUGGAAGUUGCACGUUGGG-CUGGUGUGCAUGUCCGCUAGACGGAAAGCCUUAAAGCCUACUGCAUCUUCCACCUUGAACUUUCGGGUUCAAGGGCUAAGUCGACAGCGGC (((.......((((((.((((.((.(((-((...(.((...((((.....))))..)))....))))))))))).))))))(((((((((....)))))))))............))).. ( -46.30) >ppk_1 29 107 + 1 CGCACAACCACGGGGGUUGCACGCUGGGGCCGGUGUGCAUGUCCGCCCGACGGAAAGCCUUAACGCCCCCUGCAAUCUCCACCUUGAACUUUCGGGUUCAAGGGCUA------------- .((........((((((((((....((((((((.(((......))))))..((....)).....))))).)))))))))).(((((((((....)))))))))))..------------- ( -47.20) >consensus CGCACAACCACGGAGGUUGCACGUUGGG_CCGGUGUGCAUGUCCGCACGACGGAAAGCCUUAAAGCCUACUGCAACCUCCACCUUGAACUUUCGGGUUCAAGGGCUAA_CCGACAGCGG_ ...........((((((((((....(((.((...(.((...((((.....))))..)))....)))))..)))))))))).(((((((((....)))))))))................. (-37.32 = -36.72 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:53:44 2007