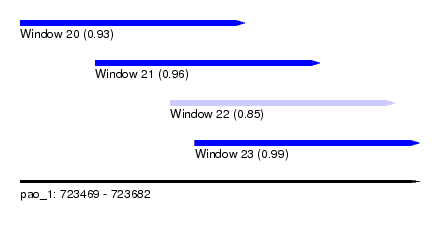
| Sequence ID | pao_1 |
|---|---|
| Location | 723,469 – 723,682 |
| Length | 213 |
| Max. P | 0.993108 |

| Location | 723,469 – 723,589 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -49.30 |
| Consensus MFE | -47.42 |
| Energy contribution | -49.30 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723469 120 + 1 UUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUA ....(((((..((....))..)))))((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))....... ( -50.10) >pf5_1 1281 120 + 1 UUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUA ....(((((..((....))..)))))((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))....... ( -48.80) >psp_1 1281 120 + 1 UUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUA ....(((((..((....))..)))))((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))....... ( -49.50) >pss_1 1281 120 + 1 UUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUA ....(((((..((....))..)))))((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))....... ( -48.80) >consensus UUCCCGGGCCUUGUACACACCGCCCGUCACACCAUGGGAGUGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUA ....(((((..((....))..)))))((((.((.(((..((((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))))..)))))))))....... (-47.42 = -49.30 + 1.88)

| Location | 723,509 – 723,629 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -56.20 |
| Consensus MFE | -54.33 |
| Energy contribution | -56.20 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.02 |
| Mean z-score | -4.37 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723509 120 + 1 UGGGUUGCUCCAGAAGUAGCUAGUCUAACCGCAAGGGGGACGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCC (((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))).....(((((((.((....))....((((((((((....))))))))))..))))))). ( -57.00) >pf5_1 1321 120 + 1 UGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCC (((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))).....(((((((.((....))....((((((((((....))))))))))..))))))). ( -55.70) >psp_1 1321 120 + 1 UGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGGGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCC (((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))).....(((((((.((....))....((((((((((....))))))))))..))))))). ( -56.40) >pss_1 1321 120 + 1 UGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCC (((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))).....(((((((.((....))....((((((((((....))))))))))..))))))). ( -55.70) >consensus UGGGUUGCACCAGAAGUAGCUAGUCUAACCUUCGGGAGGACGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCC (((((..((((.(..((((((.((((..((....)).))))))))))..)))))..))))).....(((((((.((....))....((((((((((....))))))))))..))))))). (-54.33 = -56.20 + 1.87)

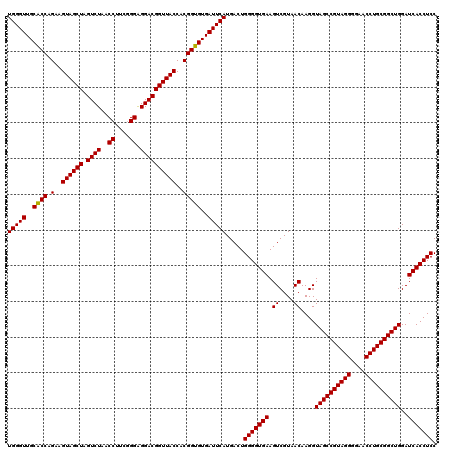
| Location | 723,549 – 723,669 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -43.28 |
| Consensus MFE | -40.78 |
| Energy contribution | -40.77 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723549 120 + 1 CGGUUACCACGGAGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGA .(((((....((((((((((((((((.......))))))........(((((((((....))))))))))))))).)))).)))))(((((........)))))................ ( -44.40) >pf5_1 1361 120 + 1 CGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCUUCAUAAGCUCCCACACGA ((.......))(((((............((((..((((....((((.(((((((((....)))))))))((....)).))))...))))..))))((((........))))..))))).. ( -43.00) >psp_1 1361 119 + 1 CGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCACCAUAAGCACCCACACGA ..........(((((.....(((..((.(((..((((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)..))).)).)))..)))))....... ( -42.70) >pss_1 1361 119 + 1 CGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCGCCAUAAGCACCCACACGA ((.......))(((((..........(.(((..((((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)..))).)((.....))...))))).. ( -43.00) >consensus CGGUUACCACGGUGUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC_UCAGCUGCUCCAUAAGCACCCACACGA ((((((....((.(((((((((((((.......))))))........(((((((((....))))))))))))))))))...)))))).........(((........))).......... (-40.78 = -40.77 + -0.00)

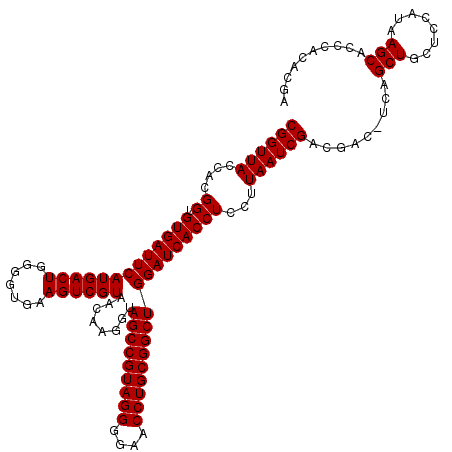
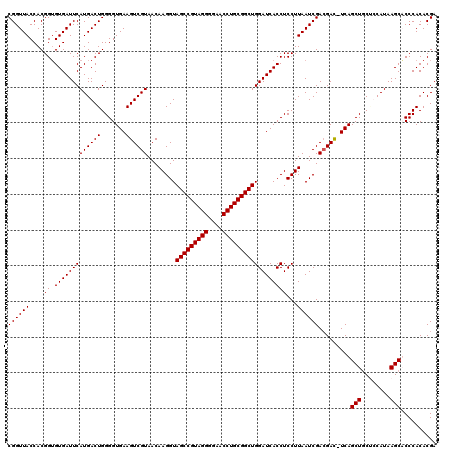
| Location | 723,562 – 723,682 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -45.25 |
| Consensus MFE | -44.71 |
| Energy contribution | -44.77 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>pao_1 723562 120 + 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGAAGAUCUCAGCUUCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((...(((((((.((....))....((((((((((....))))))))))..))))))).......(((((........)))))...........)).))))..)........ ( -41.80) >pf5_1 1374 120 + 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGACAUCAGCUGCUUCAUAAGCUCCCACACGAAUUGCUUGAUUCA (..((((.((..((((((((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........)))))))))).))))..)........ ( -46.40) >psp_1 1374 119 + 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCACCAUAAGCACCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).)))))).))))..)........ ( -46.40) >pss_1 1374 119 + 1 GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC-UCAGCUGCGCCAUAAGCACCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....)))))-)))(((........))).)))))).))))..)........ ( -46.40) >consensus GUGAUUCAUGACUGGGGUGAAGUCGUAACAAGGUAGCCGUAGGGGAACCUGCGGCUGGAUCACCUCCUUAAUCGACGAC_UCAGCUGCUCCAUAAGCACCCACACGAAUUGCUUGAUUCA (..((((.((..((((.(((.(((((...((((.(((((((((....)))))))))((....)).)))).....))))).)))(((........))).)))))).))))..)........ (-44.71 = -44.77 + 0.06)

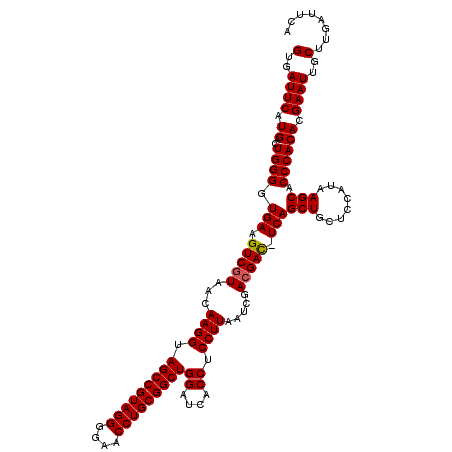
Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Jan 18 11:45:56 2007