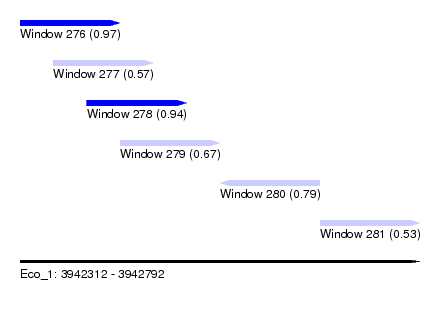
| Sequence ID | Eco_1 |
|---|---|
| Location | 3,942,312 – 3,942,792 |
| Length | 480 |
| Max. P | 0.973633 |

| Location | 3,942,312 – 3,942,432 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.81 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -41.45 |
| Energy contribution | -41.08 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942312 120 + 1 ACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGAAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGA ..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))..(((.....))).(((((.(((...)))))))).. ( -41.60) >Sfl_1 599 120 + 1 ACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGAAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGA ..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))..(((.....))).(((((.(((...)))))))).. ( -41.60) >Sty_1 601 120 + 1 ACUUAUAUUCUGUAGCAAGGUUAACCGUAUAGGGGAGCCGGAGGGAAACCGAGUCUUAAUUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGA ..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))..(((.....))).(((((.(((...)))))))).. ( -41.60) >Ype_1 598 120 + 1 ACUUAUAUUUUGUAGCAAGGUUAACCGAAUAGGGGAGCCGUAGGGAAACCGAGUCUUAACUGGGCGUCUAGUUGCAAGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGA ..((((((((((((((..((((..((.....))..))))...((....))((((((.....)))).))..))))))))))))))..(((.....))).(((((.(((...)))))))).. ( -39.80) >consensus ACUUAUAUUCUGUAGCAAGGUUAACCGAAUAGGGGAGCCGAAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGA ..((((((((((((((..((((..((.....))..))))...((....))..((((.....)))).....))))))))))))))..(((.....))).(((((.(((...)))))))).. (-41.45 = -41.08 + -0.38)

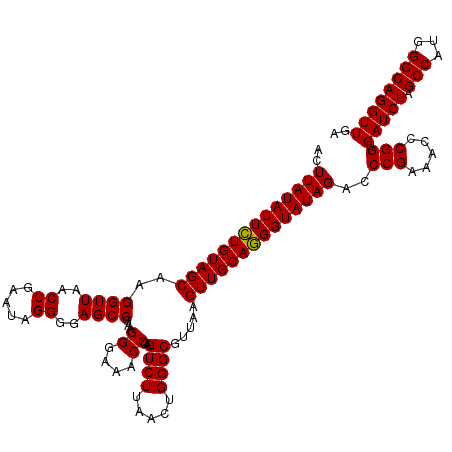
| Location | 3,942,352 – 3,942,472 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -37.61 |
| Energy contribution | -37.67 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942352 120 + 1 AAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUG ..((....)).((((...((((((.((......)).((((....))))....))))))(((((.(((...))))))))...((((.(((....(.....)....))).)))))))).... ( -39.90) >Sfl_1 639 120 + 1 AAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUG ..((....)).((((...((((((.((......)).((((....))))....))))))(((((.(((...))))))))...((((.(((....(.....)....))).)))))))).... ( -39.90) >Sty_1 641 120 + 1 GAGGGAAACCGAGUCUUAAUUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUG ..((....)).((((...((((((.((......)).((((....))))....))))))(((((.(((...))))))))...((((.(((....(.....)....))).)))))))).... ( -37.50) >Ype_1 638 120 + 1 UAGGGAAACCGAGUCUUAACUGGGCGUCUAGUUGCAAGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUG ..((....)).((((.....(((.(.((((((((....((....((((((...((..((((((.(((...)))))))))..)))))))).))..)))))))).).)))....)))).... ( -38.30) >consensus AAGGGAAACCGAGUCUUAACUGGGCGUUAAGUUGCAGGGUAUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUG ..((....)).((((...((((((.((......)).((((....))))....))))))(((((.(((...))))))))...((((.(((....(.....)....))).)))))))).... (-37.61 = -37.67 + 0.06)

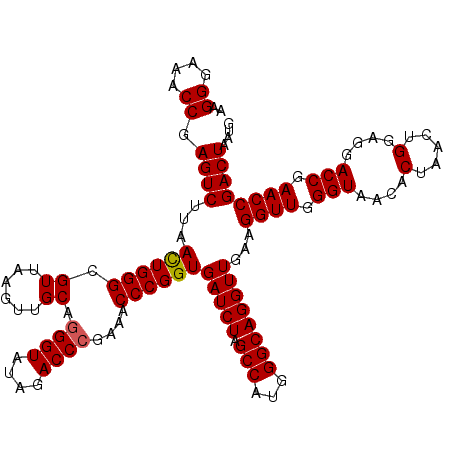
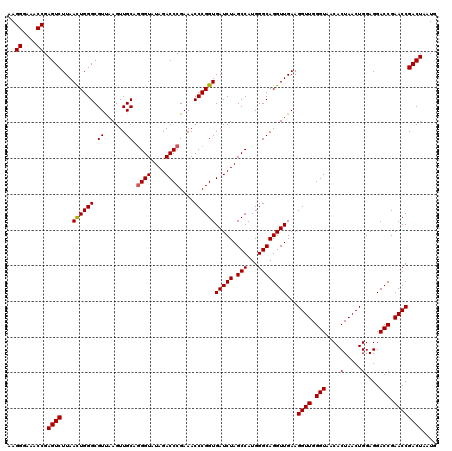
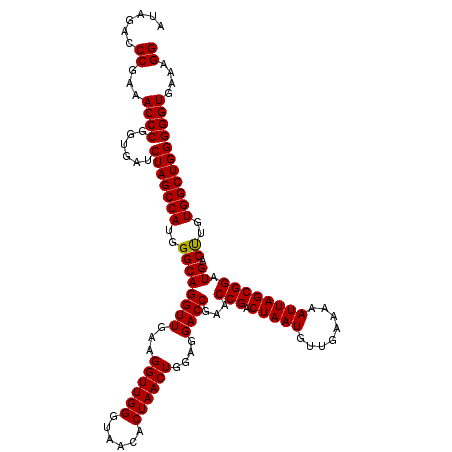
| Location | 3,942,392 – 3,942,512 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -42.84 |
| Energy contribution | -42.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942392 120 + 1 AUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGG ......((...((((......(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))))))....)) ( -42.20) >Sfl_1 679 120 + 1 AUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGG ......((...((((......(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))))))....)) ( -42.20) >Sty_1 681 120 + 1 AUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACCUGUGGCUGGGGGUGAAAGG ......((...((((......(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)))))))))))))))....)) ( -44.10) >Ype_1 678 120 + 1 AUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGG ......((...((((......(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))))))....)) ( -42.20) >consensus AUAGACCCGAAACCCGGUGAUCUAGCCAUGGGCAGGUUGAAGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGG ......((...((((......(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))))))....)) (-42.84 = -42.65 + -0.19)

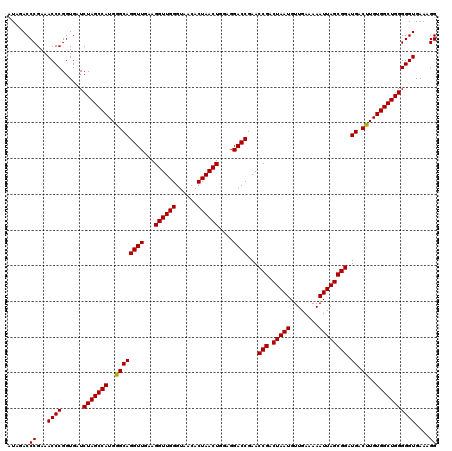
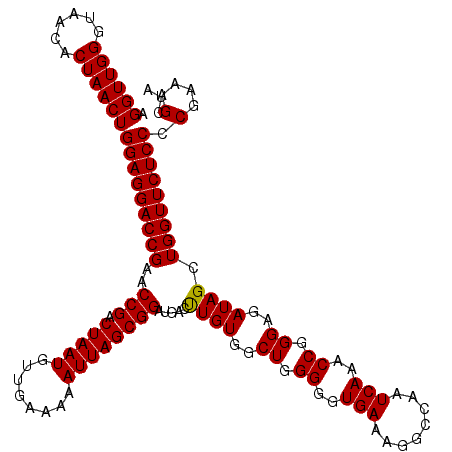
| Location | 3,942,432 – 3,942,552 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -38.71 |
| Energy contribution | -38.52 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942432 120 + 1 AGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUAGCUGGUUCUCCCCGAAAGCUA .((((((......))))))(((((((((..(((.(((((........)))))))).....((((..((.((..(((........)))..)).))..)))).))))))))).(....)... ( -38.00) >Sfl_1 719 120 + 1 AGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUAGCUGGUUCUCCCCGAAAGCUA .((((((......))))))(((((((((..(((.(((((........)))))))).....((((..((.((..(((........)))..)).))..)))).))))))))).(....)... ( -38.00) >Sty_1 721 120 + 1 AGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACCUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUAGCUGGUUCUCCCCGAAAGCUA .((((((......))))))(((((((((..(((.(((((........)))))))).....((((..((.((..(((........)))..)).))..)))).))))))))).(....)... ( -40.10) >Ype_1 718 120 + 1 AGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUAGCUGGUUCUCCCCGAAAGCUA .((((((......))))))(((((((((..(((.(((((........)))))))).....((((..((.((..(((........)))..)).))..)))).))))))))).(....)... ( -38.00) >consensus AGGUUGGGUAACACUAACUGGAGGACCGAACCGACUAAUGUUGAAAAAUUAGCGGAUGACUUGUGGCUGGGGGUGAAAGGCCAAUCAAACCGGGAGAUAGCUGGUUCUCCCCGAAAGCUA .((((((......))))))(((((((((..(((.(((((........)))))))).....((((..((.((..(((........)))..)).))..)))).))))))))).(....)... (-38.71 = -38.52 + -0.19)

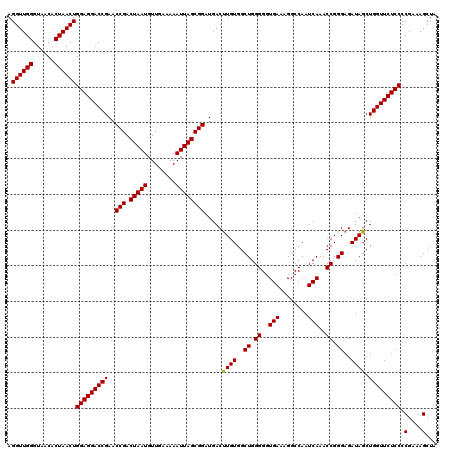
| Location | 3,942,552 – 3,942,672 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -41.58 |
| Consensus MFE | -40.62 |
| Energy contribution | -40.75 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942552 120 - 1 UGUCUCCCGUGAUAACAUUCUCCGGUAUUCGCAGUUUGCAUCGGGUUGGUAAGUCGGGAUGACCCCCUUGCCGAAACAGUGCUCUACCCCCGGAGAUGAAUUCACGAGGCGCUACCUAAA .(((((..((((...(((.((((((.....(.((...((((.(..((((((((..(((....))).))))))))..).)))).)).)..)))))))))...))))))))).......... ( -43.00) >Sfl_1 839 120 - 1 UGUCUCCCGUGAUAACAUUCUCCGGUAUUCGCAGUUUGCAUCGGGUUGGUAAGUCGGGAUGACCCCCUUGCCGAAACAGUGCUCUACCCCCGGAGAUGAAUUCACGAGGCGCUACCUAAA .(((((..((((...(((.((((((.....(.((...((((.(..((((((((..(((....))).))))))))..).)))).)).)..)))))))))...))))))))).......... ( -43.00) >Sty_1 841 120 - 1 UGUCUCCCGUGAUAACAUUCUCCGGUAUUCGCAGUUUGCAUCGGGUUGGUAAGCCGGGAUGGCCCCCUAGCCGAAACAGUGCUCUACCCCCGGAGAUGAAUUCACGAGGCGCUACCUAAA .(((((..((((...(((.((((((.....(.((...((((.(..(((((.((..(((....))).)).)))))..).)))).)).)..)))))))))...))))))))).......... ( -39.00) >Ype_1 838 120 - 1 UGUCUCCCGUGAUAACAUUCUUCGGUAUUCGGAGUUUGCAUCGGUUUGGUAAGCCGGGAUGGCCCCCUAGCCGAAACAGUGCUCUACCCCCGAAGAUGAGUUCACGAGGCGCUACCUAAA .(((((..((((...(((.((((((.....((((((((..((((((.((...(((.....)))..)).))))))..))).)))))....)))))))))...))))))))).......... ( -41.30) >consensus UGUCUCCCGUGAUAACAUUCUCCGGUAUUCGCAGUUUGCAUCGGGUUGGUAAGCCGGGAUGACCCCCUAGCCGAAACAGUGCUCUACCCCCGGAGAUGAAUUCACGAGGCGCUACCUAAA .(((((..((((...(((.((((((.......((...((((.(..((((((((..(((....))).))))))))..).)))).))....)))))))))...))))))))).......... (-40.62 = -40.75 + 0.12)


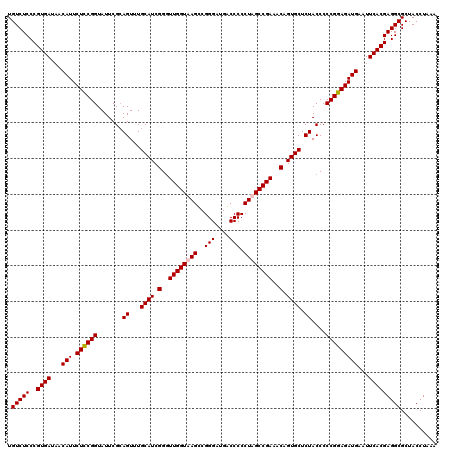
| Location | 3,942,672 – 3,942,792 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -43.12 |
| Energy contribution | -43.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.98 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 3942672 120 + 1 CACGGCGGGUGCUAACGUCCGUCGUGAAGAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCCCAGACAGCCAGGAUGUUGGCU ((((((((..(....)..))))))))....(((......))).((((.........))))((((.(((.(((((..((.(....).)).((((....))))...))))).))).)))).. ( -44.30) >Sfl_1 959 120 + 1 CACGGCGGGUGCUAACGUCCGUCGUGAAGAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCCCAGACAGCCAGGAUGUUGGCU ((((((((..(....)..))))))))....(((......))).((((.........))))((((.(((.(((((..((.(....).)).((((....))))...))))).))).)))).. ( -44.30) >Sty_1 961 120 + 1 CACGGCGGGUGCUAACGUCCGUCGUGAAGAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCCCAGACAGCCAGGAUGUUGGCU ((((((((..(....)..))))))))....(((......))).((((.........))))((((.(((.(((((..((.(....).)).((((....))))...))))).))).)))).. ( -44.30) >Ype_1 958 120 + 1 CACGGCGGGUGCUAACGUCCGUCGUGAAGAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCACAGACAGCCAGGAUGUUGGCU ((((((((..(....)..))))))))....(((......))).....((((((.....(((((....(((.(((........))).)))))))).(((.......))).....)))))). ( -43.50) >consensus CACGGCGGGUGCUAACGUCCGUCGUGAAGAGGGAAACAACCCAGACCGCCAGCUAAGGUCCCAAAGUCAUGGUUAAGUGGGAAACGAUGUGGGAAGGCCCAGACAGCCAGGAUGUUGGCU ((((((((..(....)..))))))))....(((......))).....((((((.....(((((....(((.(((........))).)))))))).(((.......))).....)))))). (-43.12 = -43.12 + 0.00)

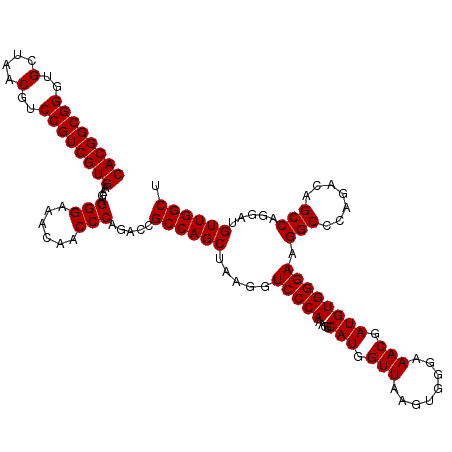
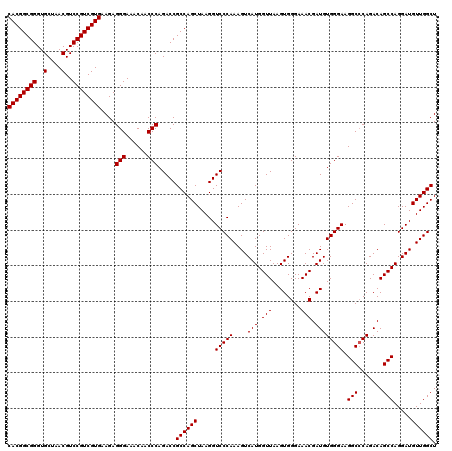
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:06:04 2006