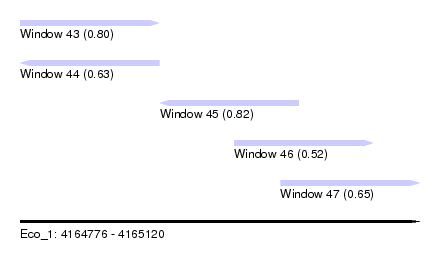
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,164,776 – 4,165,120 |
| Length | 344 |
| Max. P | 0.817212 |

| Location | 4,164,776 – 4,164,896 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -41.40 |
| Energy contribution | -41.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164776 120 + 1 AGGUGGGUGACGAAGCGGUUGCCAUGACCUGCGUUUGCAAACACCGGUACCGACAGGACAGGGUGGAGAACGAGUGAGGGGCGACUCGCAGAGUCAGUUGGACCGCAAUGUUAAGAAUAA ..((...(((((..((((((....((((((((((((.....((((.((.((....))))..))))..))))......(((....))))))).))))....))))))..)))))...)).. ( -41.40) >Sfl_1 241 120 + 1 AGGUGGGUGACGAAGCGGUUGCCAUGACCUGCGUUUGCAAACACCGGUACCGACAGGACAGGGUGGAGAACGAGUGAGGGGCGACUCGCAGAGUCAGUUGGACCGCAAUGUUAAGAAUAA ..((...(((((..((((((....((((((((((((.....((((.((.((....))))..))))..))))......(((....))))))).))))....))))))..)))))...)).. ( -41.40) >Sty_1 241 120 + 1 AGGUGGGUGACGAAGCGGUUGCCAUGACCUGCGUUUGCAAACACCGGUACCGACAGGACAGGGUGGAGAACGGGUGAGGGGCGACUCGCAGAGUCAGUUGGACCGCAAUGUUAAGAAUAA ..((...(((((..((((((....(((((((((.((((...((((.((.((.((........)).).).)).))))....))))..))))).))))....))))))..)))))...)).. ( -41.90) >consensus AGGUGGGUGACGAAGCGGUUGCCAUGACCUGCGUUUGCAAACACCGGUACCGACAGGACAGGGUGGAGAACGAGUGAGGGGCGACUCGCAGAGUCAGUUGGACCGCAAUGUUAAGAAUAA ..((...(((((..((((((....((((((((((((.....((((.((.((....))))..))))..))))......(((....))))))).))))....))))))..)))))...)).. (-41.40 = -41.40 + -0.00)

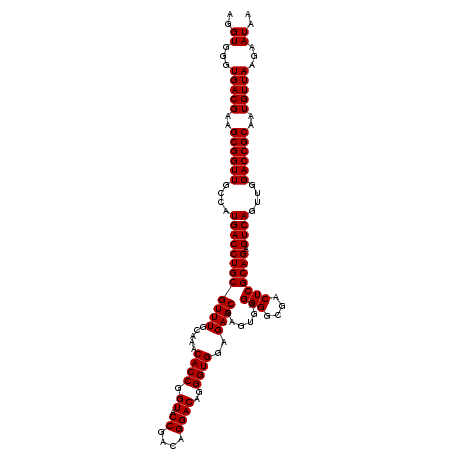
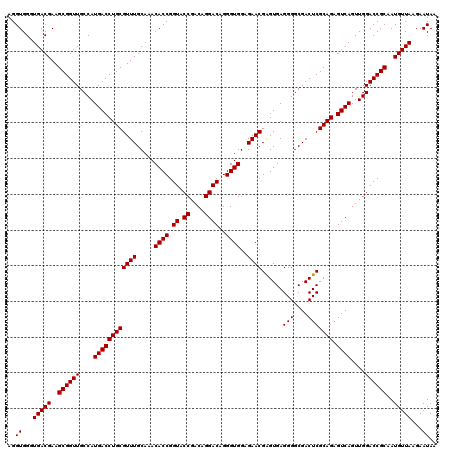
| Location | 4,164,776 – 4,164,896 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164776 120 - 1 UUAUUCUUAACAUUGCGGUCCAACUGACUCUGCGAGUCGCCCCUCACUCGUUCUCCACCCUGUCCUGUCGGUACCGGUGUUUGCAAACGCAGGUCAUGGCAACCGCUUCGUCACCCACCU ..............((((((((..((((.((((((((........))))((....((((..((((....)).)).))))...))....)))))))))))..))))).............. ( -32.30) >Sfl_1 241 120 - 1 UUAUUCUUAACAUUGCGGUCCAACUGACUCUGCGAGUCGCCCCUCACUCGUUCUCCACCCUGUCCUGUCGGUACCGGUGUUUGCAAACGCAGGUCAUGGCAACCGCUUCGUCACCCACCU ..............((((((((..((((.((((((((........))))((....((((..((((....)).)).))))...))....)))))))))))..))))).............. ( -32.30) >Sty_1 241 120 - 1 UUAUUCUUAACAUUGCGGUCCAACUGACUCUGCGAGUCGCCCCUCACCCGUUCUCCACCCUGUCCUGUCGGUACCGGUGUUUGCAAACGCAGGUCAUGGCAACCGCUUCGUCACCCACCU ..............((((((((..((((.(((((..(.((....((((.((...((((........)).)).)).))))...)))..))))))))))))..))))).............. ( -32.20) >consensus UUAUUCUUAACAUUGCGGUCCAACUGACUCUGCGAGUCGCCCCUCACUCGUUCUCCACCCUGUCCUGUCGGUACCGGUGUUUGCAAACGCAGGUCAUGGCAACCGCUUCGUCACCCACCU ..............((((((((..((((.(((((((......)))....((....((((..((((....)).)).))))...))....)))))))))))..))))).............. (-31.20 = -31.20 + -0.00)

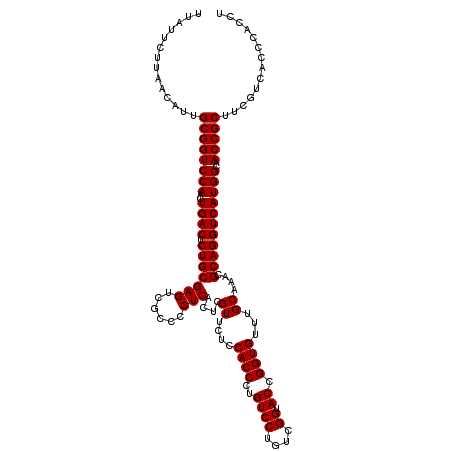
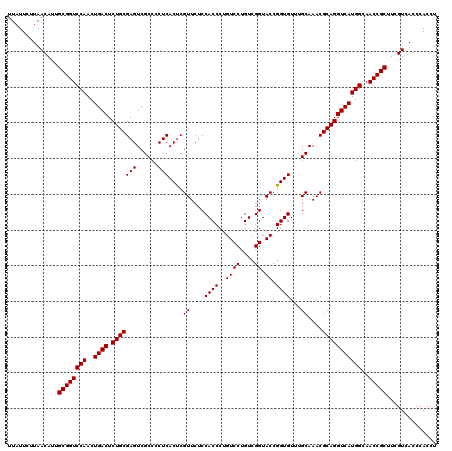
| Location | 4,164,896 – 4,165,016 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -42.83 |
| Energy contribution | -43.17 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164896 120 - 1 ACGCACUCGGGAGGACGAUCCCUUUUUCGUGGGAGCCUCUUACGAGUGCGGGGUUCCGCGGUCAAUCCAGCACGCUCUGUAUACCCCCGUACAAUUUAUCAGAGGCCCAGACAUCCCUGA .(((((((((((((....((((........)))).)))))..))))))))((((..((((((.......)).)))..(((((......)))))........)..))))............ ( -44.00) >Sfl_1 361 120 - 1 ACGCACUCGGGAGGACGAUCCCUUUUUCGUGGGAGCCUCUUACGAGUGCGGGGUUCCGCGGUCAAUCCAGCACGCUCUGUAUACCCCCGUACAAUUUAUCAGAGGCCCAGACAUCCCUGA .(((((((((((((....((((........)))).)))))..))))))))((((..((((((.......)).)))..(((((......)))))........)..))))............ ( -44.00) >Sty_1 361 120 - 1 ACGCACUCGGGAGGACGAUCCCUUUUUCGUGGGAGCCUCUUACGAGUGCGGGGUUCCGCGGACAAUCCAGCACGCUUUGUAAACUCCCGUACAAUUUAUCAGAGGCCCUGACAUCCCUGA .(((((((((((((....((((........)))).)))))..))))))))((((..((((..(......)..))).(((((........))))).......)..))))............ ( -43.30) >consensus ACGCACUCGGGAGGACGAUCCCUUUUUCGUGGGAGCCUCUUACGAGUGCGGGGUUCCGCGGUCAAUCCAGCACGCUCUGUAUACCCCCGUACAAUUUAUCAGAGGCCCAGACAUCCCUGA .(((((((((((((....((((........)))).)))))..))))))))((((..((((..(......)..)))..(((((......)))))........)..))))............ (-42.83 = -43.17 + 0.33)

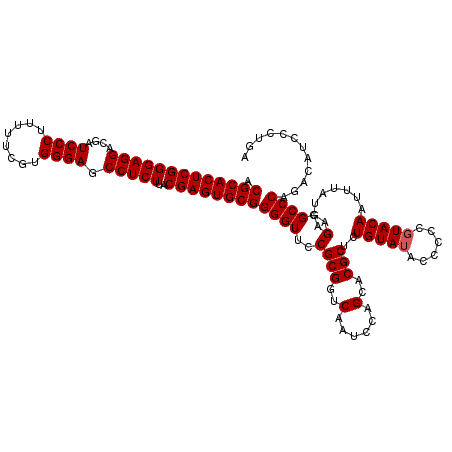
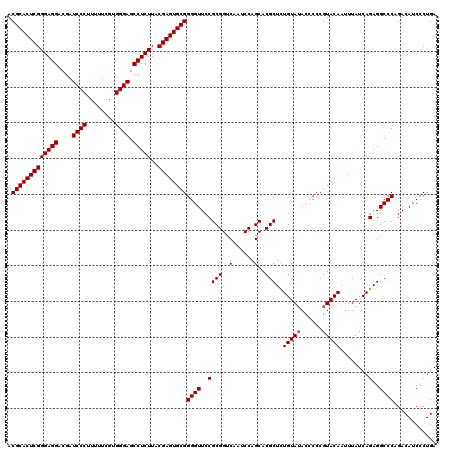
| Location | 4,164,960 – 4,165,080 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -46.03 |
| Consensus MFE | -40.45 |
| Energy contribution | -41.23 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520104 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4164960 120 + 1 CGGAGGCUAUAUCGCGGUGGAUACCCGACAAAGAACAGACAUUUCGGCAUAUAUGCCAGAGCUACCAAUGGCAGGGCCGCGGUAUCACCGCGGCAUGGCGAACGGUACUGUAGCGAAGCC ....((((...((((((.(....)))........((((((..((((.(((...(((((..........)))))..(((((((.....)))))))))).))))..)).)))).)))))))) ( -47.10) >Hin_1 278 120 + 1 GGGAGGGUAUAUCGCGGUGGAUACCCGACAAAGAACAGACAUUUCGGCAUAUAUGCCAGAGCUACCAAUGGCAGGGCCGCGGUAUCACCGCGCCAUGGCGGAUGGCACUGUGGCGAAGCC .(..((((((.((.....))))))))..)................(((.....(((((..........)))))..((((((((..((((((......)))).))..))))))))...))) ( -43.90) >Ype_1 281 120 + 1 CGGAGGCUAUAUCGCGGUGGAUACCCGACAAAGAACAGACAUUUCGGCAUAUAUGCCAGAGCUACCAAUGGCAGGGCCGCGGUAUCACCGCGGCAUGGCGAACGGUACUGUAGCGAAGCC ....((((...((((((.(....)))........((((((..((((.(((...(((((..........)))))..(((((((.....)))))))))).))))..)).)))).)))))))) ( -47.10) >consensus CGGAGGCUAUAUCGCGGUGGAUACCCGACAAAGAACAGACAUUUCGGCAUAUAUGCCAGAGCUACCAAUGGCAGGGCCGCGGUAUCACCGCGGCAUGGCGAACGGUACUGUAGCGAAGCC ....((((...((((((.(....)))........((((((..((((.(((...(((((..........)))))..(((((((.....)))))))))).))))..)).)))).)))))))) (-40.45 = -41.23 + 0.78)

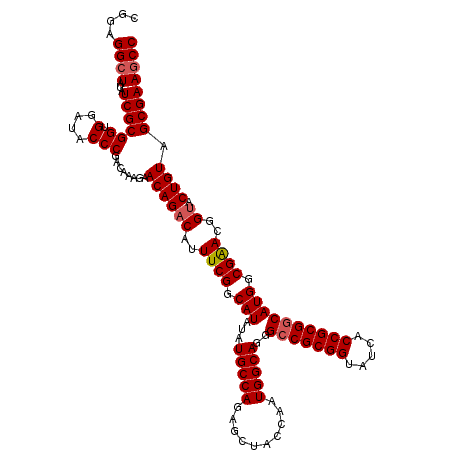
| Location | 4,165,000 – 4,165,120 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -41.01 |
| Energy contribution | -41.23 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4165000 120 + 1 AGGGACUGAAGCAAAUGCUAUCCUGUCUAGGCCCCGGAGACCAUCAGGUUGGACGAAACGUGUACCUACGUUUGGGGAUAGAGGGGAAACAGCUGUGGGCUGGGAUACCCGAUCCAGUCA ...(((((.(((..(((((.((((.((((..(((((..((((....))))...).(((((((....))))))))))).)))).))))...))).))..)))(((...)))....))))). ( -44.90) >Hin_1 318 120 + 1 AGGGACUGAAGCAAAUGCUAUCCUGUCUAGGCCCCGGAGACCAUCAGGUUGGGCGAAACGUGAACGUACGUCUGGGGAUAGAGGGGAAUGAGCUGUGGGCUGGGAUACCCGAUCCACUCA ................(((.((((.((((..((((...((((....))))(((((..(((....))).))))))))).)))).))))...))).((((..((((...))))..))))... ( -46.70) >Ype_1 321 120 + 1 AGGGACUGAAGCAAAUGCUAUCCUGUCUAGGCCCCGGAGACCAUCAGGUUGGACGAAACGCGUACCUACGUUUGGGGAUAGAGGGGAAACAGCUGUGGGCUGGGAUACCCGAUCCGGUCA ...(((((.(((..(((((.((((.((((..(((((((((((....))))(((((.....))).))....))))))).)))).))))...))).))..)))(((...)))....))))). ( -44.00) >consensus AGGGACUGAAGCAAAUGCUAUCCUGUCUAGGCCCCGGAGACCAUCAGGUUGGACGAAACGUGUACCUACGUUUGGGGAUAGAGGGGAAACAGCUGUGGGCUGGGAUACCCGAUCCAGUCA ...(((((.(((..(((((.((((.((((..(((((((((((....))))........((((....))))))))))).)))).))))...))).))..)))(((...)))....))))). (-41.01 = -41.23 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Thu Dec 7 14:12:20 2006