
| Sequence ID | Eco_1 |
|---|---|
| Location | 4,178,949 – 4,179,268 |
| Length | 319 |
| Max. P | 0.992563 |

| Location | 4,178,949 – 4,179,069 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -56.70 |
| Consensus MFE | -54.70 |
| Energy contribution | -56.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.76 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4178949 120 + 1 GCCAACCCUUCCGGUUGCAGCCUGAGAAAUCAGGCUGAUGGCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCC (((((((.....)))))((((((((....))))))))..(((((((((((....))))))))))).....)).......(((((((((((((...(.....)..)).))))))..))))) ( -56.70) >Sfl_1 1 120 + 1 GCCAACCCUUCCGGUUGCAGCCUGAGAAAUCAGGCUGAUGGCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCC (((((((.....)))))((((((((....))))))))..(((((((((((....))))))))))).....)).......(((((((((((((...(.....)..)).))))))..))))) ( -56.70) >Sty_1 1 120 + 1 GCCAACCCUUCCGGUUGCAGCCUGAGAAAUCAGGCUGAUGGCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCC (((((((.....)))))((((((((....))))))))..(((((((((((....))))))))))).....)).......(((((((((((((...(.....)..)).))))))..))))) ( -56.70) >consensus GCCAACCCUUCCGGUUGCAGCCUGAGAAAUCAGGCUGAUGGCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCC (((((((.....)))))((((((((....))))))))..(((((((((((....))))))))))).....)).......(((((((((((((...(.....)..)).))))))..))))) (-54.70 = -56.70 + 2.00)

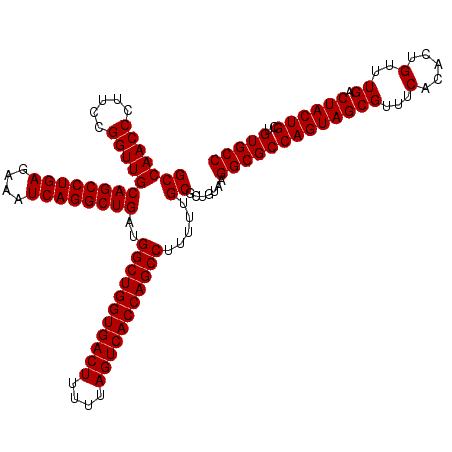
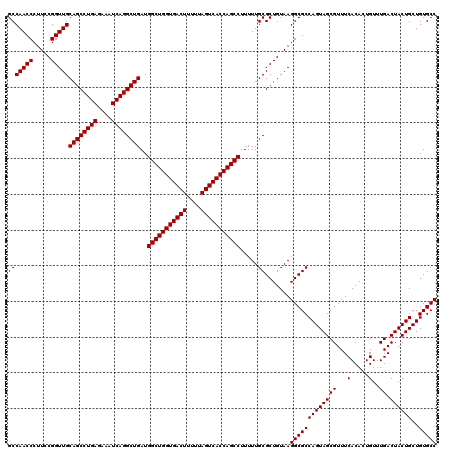
| Location | 4,178,949 – 4,179,069 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -55.20 |
| Consensus MFE | -55.20 |
| Energy contribution | -55.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -5.73 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906145 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4178949 120 - 1 GGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGCCAUCAGCCUGAUUUCUCAGGCUGCAACCGGAAGGGUUGGC (((.(((.(((..(((.......)))...))).)))..)))...((((((.....(((((((((((....)))))))))))...(((((((....)))))))))..((....)))))).. ( -55.20) >Sfl_1 1 120 - 1 GGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGCCAUCAGCCUGAUUUCUCAGGCUGCAACCGGAAGGGUUGGC (((.(((.(((..(((.......)))...))).)))..)))...((((((.....(((((((((((....)))))))))))...(((((((....)))))))))..((....)))))).. ( -55.20) >Sty_1 1 120 - 1 GGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGCCAUCAGCCUGAUUUCUCAGGCUGCAACCGGAAGGGUUGGC (((.(((.(((..(((.......)))...))).)))..)))...((((((.....(((((((((((....)))))))))))...(((((((....)))))))))..((....)))))).. ( -55.20) >consensus GGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGCCAUCAGCCUGAUUUCUCAGGCUGCAACCGGAAGGGUUGGC (((.(((.(((..(((.......)))...))).)))..)))...((((((.....(((((((((((....)))))))))))...(((((((....)))))))))..((....)))))).. (-55.20 = -55.20 + -0.00)

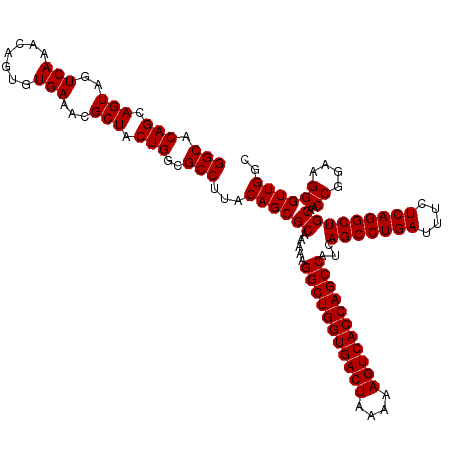
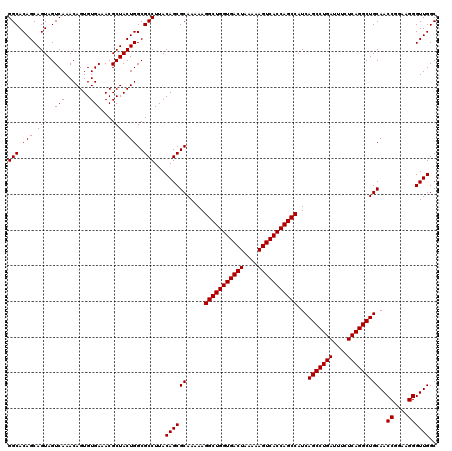
| Location | 4,178,989 – 4,179,109 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -40.60 |
| Energy contribution | -40.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.80 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
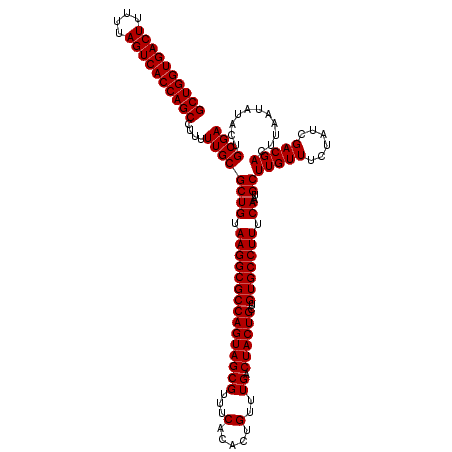
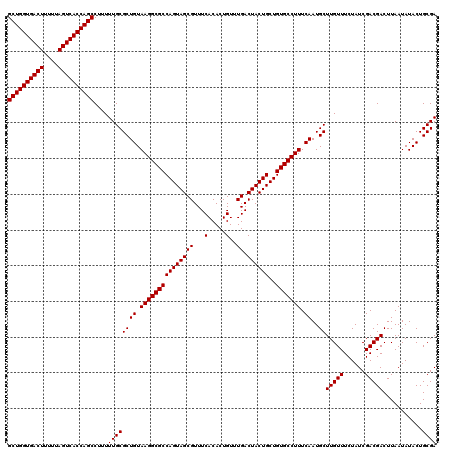
>Eco_1 4178989 120 + 1 GCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCCUUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGA ((((((((((....))))))))))....((((((((.(((((((((((((((...(.....)..)).))))))..))))))).))..))(((((......)))))...........)))) ( -40.60) >Sfl_1 41 120 + 1 GCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCCUUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGA ((((((((((....))))))))))....((((((((.(((((((((((((((...(.....)..)).))))))..))))))).))..))(((((......)))))...........)))) ( -40.60) >Sty_1 41 120 + 1 GCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCCUUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGA ((((((((((....))))))))))....((((((((.(((((((((((((((...(.....)..)).))))))..))))))).))..))(((((......)))))...........)))) ( -40.60) >consensus GCUGGUGACUUUUUAGUCACCAGCCUUUUUGCGCUGUAAGGCGCCAGUAGCGUUUCACACUGUUUGACUACUGCUGUGCCUUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGA ((((((((((....))))))))))....((((((((.(((((((((((((((...(.....)..)).))))))..))))))).))..))(((((......)))))...........)))) (-40.60 = -40.60 + 0.00)

| Location | 4,178,989 – 4,179,109 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -38.20 |
| Energy contribution | -38.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
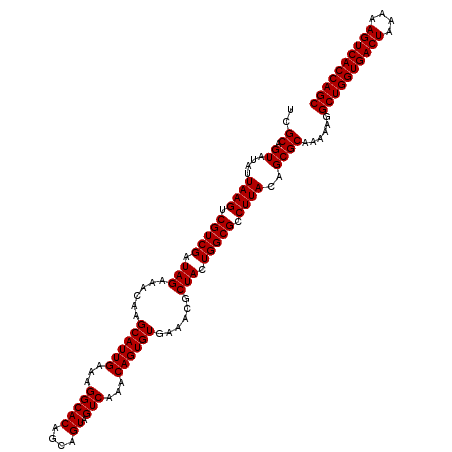
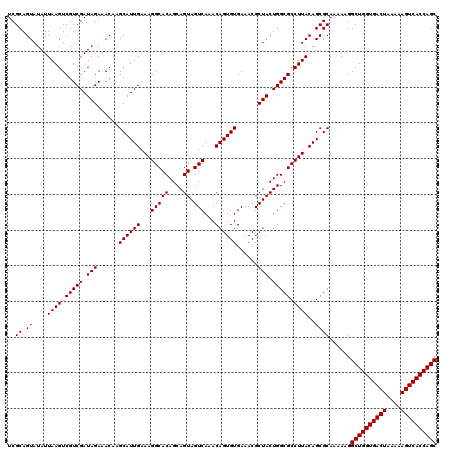
>Eco_1 4178989 120 - 1 UCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAAGGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGC ..((.((....((((.(((((.(((......((((((...(((((....)).)))...))))))......))).))))).))))..))))......((((((((((....)))))))))) ( -38.20) >Sfl_1 41 120 - 1 UCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAAGGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGC ..((.((....((((.(((((.(((......((((((...(((((....)).)))...))))))......))).))))).))))..))))......((((((((((....)))))))))) ( -38.20) >Sty_1 41 120 - 1 UCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAAGGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGC ..((.((....((((.(((((.(((......((((((...(((((....)).)))...))))))......))).))))).))))..))))......((((((((((....)))))))))) ( -38.20) >consensus UCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAAGGCACAGCAGUAGUCAAACAGUGUGAAACGCUACUGGCGCCUUACAGCGCAAAAAGGCUGGUGACUAAAAAGUCACCAGC ..((.((....((((.(((((.(((......((((((...(((((....)).)))...))))))......))).))))).))))..))))......((((((((((....)))))))))) (-38.20 = -38.20 + -0.00)

| Location | 4,179,069 – 4,179,189 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -39.53 |
| Consensus MFE | -38.83 |
| Energy contribution | -38.73 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4179069 120 + 1 UUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGACAGGACGUCCGUUCUGUGUAAAUCGCAAUGAAAUGGUUUAAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUU ...(((((((((((.(((((.(((.(((((.....(((((((((((....))))))......)))))..........))))))))))))).))))))))))).((((....))))..... ( -39.66) >Sfl_1 121 120 + 1 UUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGACAGGACGUCCGUUCUGUGUAAAUCGCAAUGAAAUGGUUUAAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUU ...(((((((((((.(((((.(((.(((((.....(((((((((((....))))))......)))))..........))))))))))))).))))))))))).((((....))))..... ( -39.66) >Sty_1 121 119 + 1 UUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGACAGAGCGUACGUUCUGUGUAAAUCGCAAUGAAAUGGUUUAAGCGUGAUAGCAACAGGCAUUGCGGAAAGUAU-CCAUUUU ...(((((((((((.(((((.(((.(((((.....(((((((((((....))))))......)))))..........))))))))))))).))))))))))).(((.....)-))..... ( -39.26) >consensus UUUCAAUGCUUGUUUCUAUCGACGACUUAAUAUACUGCGACAGGACGUCCGUUCUGUGUAAAUCGCAAUGAAAUGGUUUAAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUU ...(((((((((((.(((((.(((.(((((.....(((((((((((....))))))......)))))..........))))))))))))).))))))))))).((((....))))..... (-38.83 = -38.73 + -0.11)

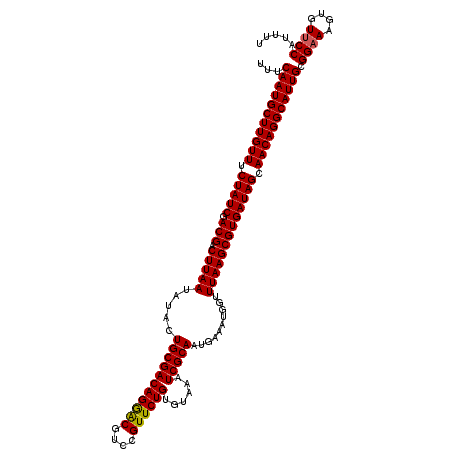
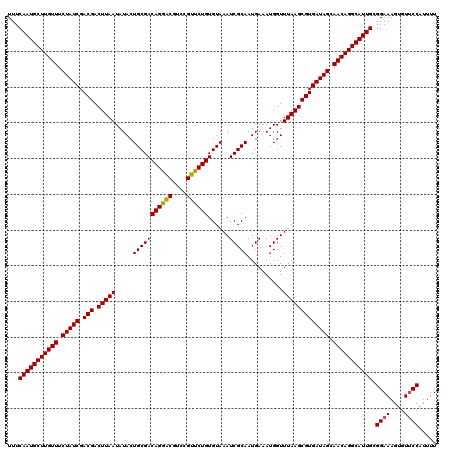
| Location | 4,179,069 – 4,179,189 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.47 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4179069 120 - 1 AAAAUGGAACACUUUCCGCAAUGCCUGUUGCUAUCACGCUUAAACCAUUUCAUUGCGAUUUACACAGAACGGACGUCCUGUCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAA .....((((....)))).((((((.((((.(((((((((((((........(((((((......(((.((....)).))))))))))...))))).))).))))).)))).))))))... ( -34.00) >Sfl_1 121 120 - 1 AAAAUGGAACACUUUCCGCAAUGCCUGUUGCUAUCACGCUUAAACCAUUUCAUUGCGAUUUACACAGAACGGACGUCCUGUCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAA .....((((....)))).((((((.((((.(((((((((((((........(((((((......(((.((....)).))))))))))...))))).))).))))).)))).))))))... ( -34.00) >Sty_1 121 119 - 1 AAAAUGG-AUACUUUCCGCAAUGCCUGUUGCUAUCACGCUUAAACCAUUUCAUUGCGAUUUACACAGAACGUACGCUCUGUCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAA .....((-(.....))).((((((.((((.(((((((((((((........(((((((......((((.(....).)))))))))))...))))).))).))))).)))).))))))... ( -34.20) >consensus AAAAUGGAACACUUUCCGCAAUGCCUGUUGCUAUCACGCUUAAACCAUUUCAUUGCGAUUUACACAGAACGGACGUCCUGUCGCAGUAUAUUAAGUCGUCGAUAGAAACAAGCAUUGAAA .....((((....)))).((((((.((((.(((((((((((((........(((((((......(((.((....)).))))))))))...))))).))).))))).)))).))))))... (-31.80 = -32.47 + 0.67)

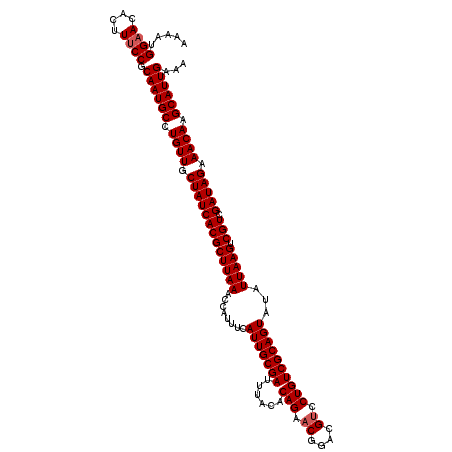
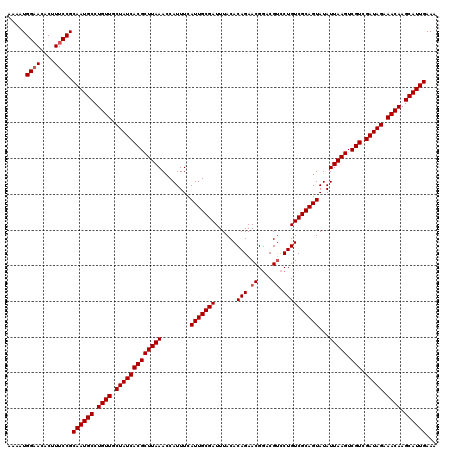
| Location | 4,179,148 – 4,179,268 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -36.27 |
| Energy contribution | -36.60 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.546187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Eco_1 4179148 120 + 1 AAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUUCCGGUCAACAAAAUAGUGUUGCACAAACUGUCCGCUCAAUGGACAGAUGGGUCGACUUGUCAGCGAGCUGAGGAACCCU ..((.((.......)).))....(((((((((...)))))))))(((((((......))))).))...(((((((.....)))))))..((((....((.((((....)))).)))))). ( -38.20) >Sfl_1 200 120 + 1 AAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUUCCGGUCAACAAAAUAGUGUUGCACAAACUGUCCGCUCAAUGGACAGAUGGGUCGACUUGUCAGCGAGCUGAGGAACCCU ..((.((.......)).))....(((((((((...)))))))))(((((((......))))).))...(((((((.....)))))))..((((....((.((((....)))).)))))). ( -38.20) >Sty_1 200 118 + 1 AAGCGUGAUAGCAACAGGCAUUGCGGAAAGUAU-CCAUUUUCCGGUCAACAAAAUAGUGUUGCACAAACUGUCC-CCCGCAGGACAGAUGGGUCGACUUGUCAGCGAGCUGAGGAACCCU ..((.((.......)).))....((((((((..-..))))))))(((((((......))))).))...((((((-......))))))..((((....((.((((....)))).)))))). ( -36.20) >consensus AAGCGUGAUAGCAACAGGCAUUGCGGAAAGUGUUCCAUUUUCCGGUCAACAAAAUAGUGUUGCACAAACUGUCCGCUCAAUGGACAGAUGGGUCGACUUGUCAGCGAGCUGAGGAACCCU ..((.((.......)).))....((((((((.....))))))))(((((((......))))).))...(((((((.....)))))))..((((....((.((((....)))).)))))). (-36.27 = -36.60 + 0.33)

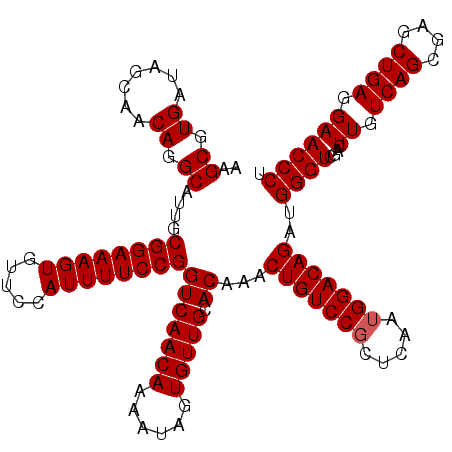
Generated by rnazCluster.pl (part of RNAz 1.0) on Fri Dec 1 11:07:37 2006