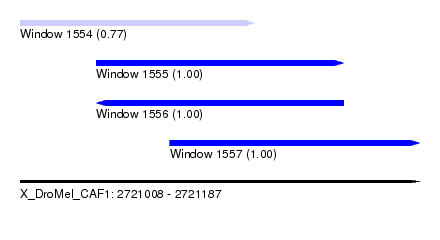
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,721,008 – 2,721,187 |
| Length | 179 |
| Max. P | 0.999986 |

| Location | 2,721,008 – 2,721,113 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.59 |
| Mean single sequence MFE | -23.78 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2721008 105 + 22224390 G-----AUGGGC-AAAAGGGGGGCGUGGCGGAUCGUAGUGCAAAUUGCGAAAGUGGAAAAU-------GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAA .-----.((.((-(..(..(((((((.((...(((((((....)))))))..)).).....-------......((((((((......))))))))..))))).)...)..))).)). ( -23.90) >DroSec_CAF1 109654 106 + 1 C-----AUGGGCCGAAAAGGGGGCGUGGCGGAUGGUAGUGCAAAUUGCGAAAGUGGAAAAU-------GGCAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAA (-----(((..((......))..))))(((.((.((.(((....(..(....)..).....-------(((((.((((((((......)))))))).))))))))..)).)).))).. ( -27.80) >DroSim_CAF1 97719 106 + 1 G-----AUGGGCCGAAAAGGGGGCGUGGCGGCUGGUAGUGCAAAUUGCGAAAGUGGAAAAU-------GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAA .-----((.(((((....(....)....))))).)).(((((..(..(....)..)....(-------((.((.((((((((......)))))))).))..))).......))))).. ( -25.10) >DroYak_CAF1 118686 108 + 1 GGGAAAAUGG----------GGUCGUGGCGGAUCGUAGUGCAAAUUGCAAAAGUGGAAAAUGGAAAAUGGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAA ..........----------.......(((.(((((..(((.....)))...((((..(((......(....).((((((((......)))))))))))..))))..))))).))).. ( -18.30) >consensus G_____AUGGGC_GAAAAGGGGGCGUGGCGGAUCGUAGUGCAAAUUGCGAAAGUGGAAAAU_______GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAA ......................(((((.((....((.(.((((.(..(....)..)..................((((((((......)))))))).)))).)))...)).))))).. (-19.17 = -19.43 + 0.25)

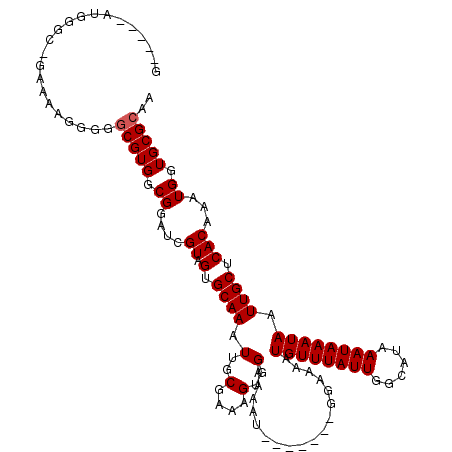
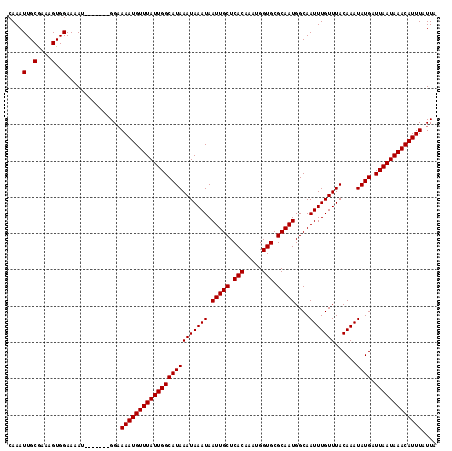
| Location | 2,721,042 – 2,721,153 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -24.92 |
| Energy contribution | -24.92 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.41 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2721042 111 + 22224390 CAAAUUGCGAAAGUGGAAAAU-------GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..).....-------...((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... ( -26.40) >DroSec_CAF1 109689 111 + 1 CAAAUUGCGAAAGUGGAAAAU-------GGCAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..).....-------...((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... ( -26.40) >DroSim_CAF1 97754 111 + 1 CAAAUUGCGAAAGUGGAAAAU-------GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..).....-------...((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... ( -26.40) >DroEre_CAF1 124147 111 + 1 CAAAUUGCAAAAGUGGAAAAU-------GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..).....-------...((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... ( -22.70) >DroYak_CAF1 118716 118 + 1 CAAAUUGCAAAAGUGGAAAAUGGAAAAUGGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..)...............((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... ( -22.70) >consensus CAAAUUGCGAAAGUGGAAAAU_______GGAAAAUGUUUAUUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUA ....(..(....)..)...............((((((((((((((((((((((((.(((((.(((.....))).)))))....)))))))......)))).))))))))))))).... (-24.92 = -24.92 + -0.00)

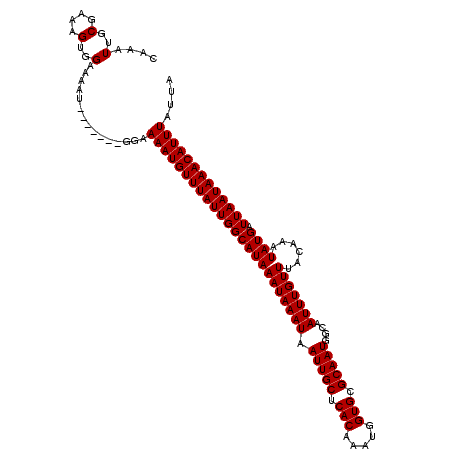
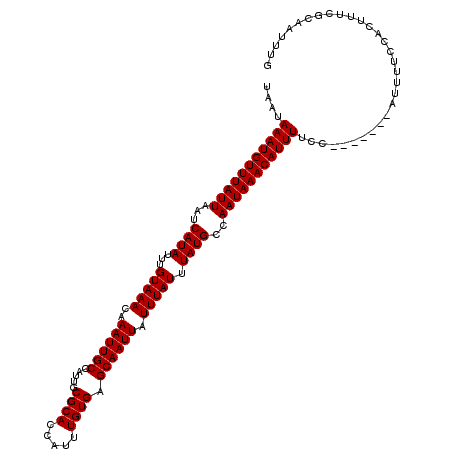
| Location | 2,721,042 – 2,721,153 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2721042 111 - 22224390 UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUUCC-------AUUUUCCACUUUCGCAAUUUG ....(((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..)))))))))))...-------..................... ( -17.90) >DroSec_CAF1 109689 111 - 1 UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUGCC-------AUUUUCCACUUUCGCAAUUUG ...((((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..))))))))))))..-------..................... ( -19.40) >DroSim_CAF1 97754 111 - 1 UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUUCC-------AUUUUCCACUUUCGCAAUUUG ....(((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..)))))))))))...-------..................... ( -17.90) >DroEre_CAF1 124147 111 - 1 UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUUCC-------AUUUUCCACUUUUGCAAUUUG ....(((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..)))))))))))...-------..................... ( -17.90) >DroYak_CAF1 118716 118 - 1 UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUUCCAUUUUCCAUUUUCCACUUUUGCAAUUUG ....(((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..)))))))))))............................... ( -17.90) >consensus UAAUAAAUGUUUAUUAAUCAUAUUUGUAAACAAAUUGCCAUUGCGCACCAUUUGUGAGCAAUUAUUUAUUUAUGCCAAUAAACAUUUUCC_______AUUUUCCACUUUCGCAAUUUG ....(((((((((((...((((...(((((..((((((.....((((.....)))).)))))).))))).))))..)))))))))))............................... (-18.02 = -18.02 + -0.00)

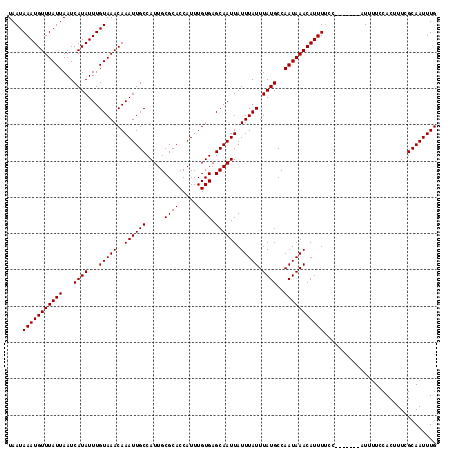
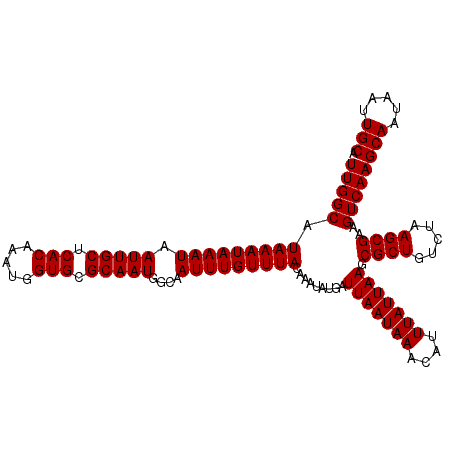
| Location | 2,721,075 – 2,721,187 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 2721075 112 + 22224390 UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). ( -25.70) >DroSec_CAF1 109722 112 + 1 UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). ( -25.70) >DroSim_CAF1 97787 112 + 1 UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). ( -25.70) >DroEre_CAF1 124180 112 + 1 UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). ( -25.70) >DroYak_CAF1 118756 112 + 1 UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). ( -25.70) >consensus UUGGCAUAAAUAAAUAAUUGCUCACAAAUGGUGCGCAAUGGCAAUUUGUUUACAAAUAUGAUUAAUAAACAUUUAUUAAGCGCUGUCUAAGCGAAGUCAAGCAAUAAUUGCA (((((.(((((((((.(((((.(((.....))).)))))....))))))))).........(((((((....))))))).((((.....))))..)))))(((.....))). (-25.70 = -25.70 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:58:07 2006