
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,994,528 – 1,994,767 |
| Length | 239 |
| Max. P | 0.976318 |

| Location | 1,994,528 – 1,994,647 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.70 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1994528 119 + 22224390 UAUUUCCCUCUUUCGCCUUUUCCUCACAAGUUCAACACAACAAGAAGUCGAGCAUGCGCUUUUCUUCUUGCCAUUUA-UAAUUGCCUUCAACUUAUUGCUCCUUGUGUUUGGGGCGAAUG ...........((((((((.....((((((..........(((((((..((((....))))..))))))).......-.....((............))..))))))...)))))))).. ( -30.10) >DroSec_CAF1 26571 120 + 1 UAUUUCCCUCUUUUGCCUUUUCCUCACAUGUUGAACUGUACAAGAAUUCGAGCAUGCGCUCUUCUUCUUGCCAUUCAUUAAUUGCCUCCAACUUAUUGCUUGUUGUUUUUGGGGCGAACG ...............................((((.((..((((((...((((....))))...)))))).))))))....(((((((((((.........)))).....)))))))... ( -22.50) >DroEre_CAF1 16312 118 + 1 UGUUUCCCUCUUUCGCCUCUUCCUCGCACGCUCAACUGUACAAGAAGCCGAGCAUGCGCUCUUCUUCUUGCCAUUCAUUAAUUGCCUCCAACUUAUUGCGCGUUGUUUUUGGGGCGCA-- .............((((((.....((.((((.(((.....(((((((..((((....))))..)))))))...........(((....)))....))).)))))).....))))))..-- ( -30.10) >DroYak_CAF1 27200 118 + 1 UAUUUCCCUCUUUCGCCUCUUCCUCGUACGCUCAACUGUACAAGAAGUCGAGCAUGCGCUCUUCUUCUUGCCAUUCAUUAAUUGCCUCCAACUUAUUGCGCGUUGAUUUUGGGGCGCA-- .............((((((....(((.((((.(((.....(((((((..((((....))))..)))))))...........(((....)))....))).)))))))....))))))..-- ( -32.10) >consensus UAUUUCCCUCUUUCGCCUCUUCCUCACACGCUCAACUGUACAAGAAGUCGAGCAUGCGCUCUUCUUCUUGCCAUUCAUUAAUUGCCUCCAACUUAUUGCGCGUUGUUUUUGGGGCGAA__ ...........((((((((.....((.((((.(((.....(((((((..((((....))))..)))))))...........(((....)))....))).)))))).....)))))))).. (-23.14 = -23.70 + 0.56)

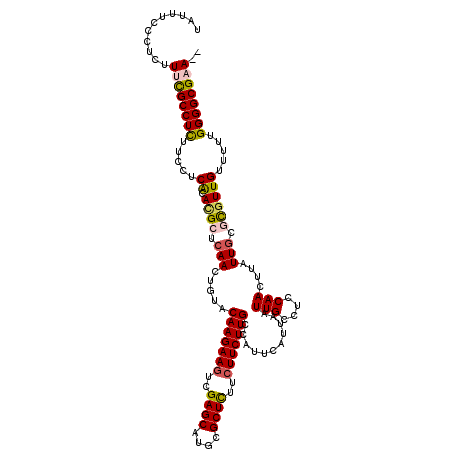
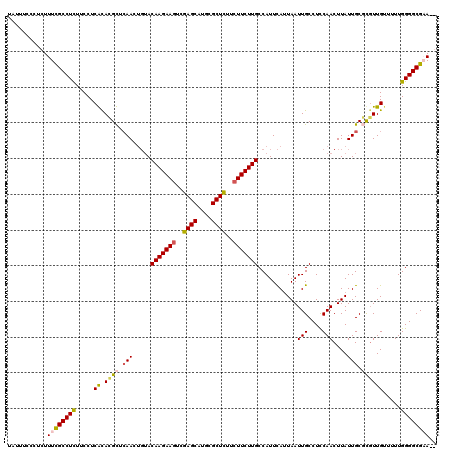
| Location | 1,994,528 – 1,994,647 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.16 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -23.52 |
| Energy contribution | -25.02 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1994528 119 - 22224390 CAUUCGCCCCAAACACAAGGAGCAAUAAGUUGAAGGCAAUUA-UAAAUGGCAAGAAGAAAAGCGCAUGCUCGACUUCUUGUUGUGUUGAACUUGUGAGGAAAAGGCGAAAGAGGGAAAUA ..((((((((...((((((...((((.(((((....))))).-...(..((((((((...(((....)))...))))))))..)))))..)))))).))....))))))........... ( -35.00) >DroSec_CAF1 26571 120 - 1 CGUUCGCCCCAAAAACAACAAGCAAUAAGUUGGAGGCAAUUAAUGAAUGGCAAGAAGAAGAGCGCAUGCUCGAAUUCUUGUACAGUUCAACAUGUGAGGAAAAGGCAAAAGAGGGAAAUA (.((((((((((.................)))).)))......(((((.(((((((...((((....))))...)))))))...))))).....................))).)..... ( -29.43) >DroEre_CAF1 16312 118 - 1 --UGCGCCCCAAAAACAACGCGCAAUAAGUUGGAGGCAAUUAAUGAAUGGCAAGAAGAAGAGCGCAUGCUCGGCUUCUUGUACAGUUGAGCGUGCGAGGAAGAGGCGAAAGAGGGAAACA --..((((((.....((((.........))))...(((.(((((.....((((((((..((((....))))..))))))))...)))))...)))..))....))))......(....). ( -36.00) >DroYak_CAF1 27200 118 - 1 --UGCGCCCCAAAAUCAACGCGCAAUAAGUUGGAGGCAAUUAAUGAAUGGCAAGAAGAAGAGCGCAUGCUCGACUUCUUGUACAGUUGAGCGUACGAGGAAGAGGCGAAAGAGGGAAAUA --..((((.(....((.((((.((((.(((((....)))))........((((((((..((((....))))..))))))))...)))).))))..))....).))))............. ( -34.50) >consensus __UGCGCCCCAAAAACAACGAGCAAUAAGUUGGAGGCAAUUAAUGAAUGGCAAGAAGAAGAGCGCAUGCUCGACUUCUUGUACAGUUGAACGUGCGAGGAAAAGGCGAAAGAGGGAAAUA .......(((.......((((.((((.(((((....)))))........((((((((..((((....))))..))))))))...)))).))))............(....).)))..... (-23.52 = -25.02 + 1.50)

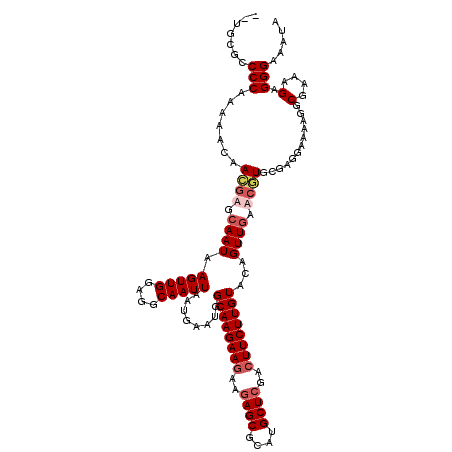
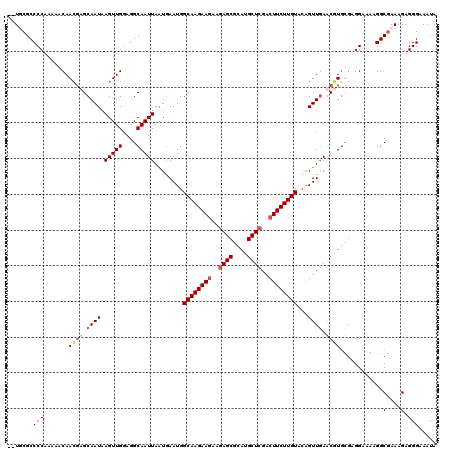
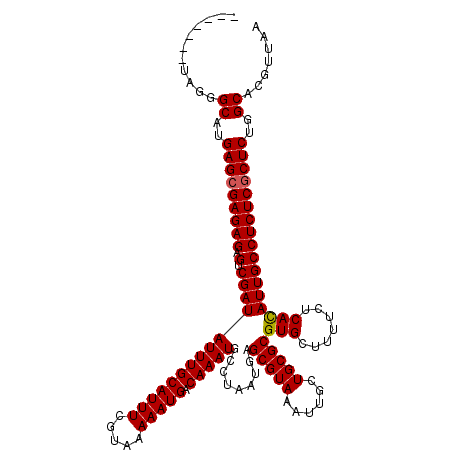
| Location | 1,994,647 – 1,994,767 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -36.16 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1994647 120 + 22224390 GUACUCCUAGGGCAUGAGCGAGAGAGUCGAUAUUUGCAUUUCGUAAAAAUGACAAAUGCCUAAUGAGCGUAAAUUGCUGCGCGUGCUUUUGUCAUAUUGCCUCUCACUCUGGCACGUUAA ..........(.((.(((.(((((.(.((((((((((.....)))))..(((((((.((.......(((((......)))))..)).)))))))))))))))))).))))).)....... ( -33.80) >DroSec_CAF1 26691 120 + 1 AUACUCCUAGGGCAUGAGCGAGAGAGUCGAUAUUUGCAUUUCGUAAAAAUGACAAAUGUCUAAUGAGCGUAAAUUGCUGCGCGUGCUUUUCUCACAUUGCCUCUCGCUCUAGCACGUUAA ...........((..(((((((((.((.(((((((((((((.....))))).))))))))......(((((......)))))(((.......)))...)))))))))))..))....... ( -37.50) >DroSim_CAF1 20891 120 + 1 NNNNNNNNNGGGCAUGAGCGAGAGAGUCGAUAUUUGCAUUUCGUAAAAAUGACAAAUGCCUAAUGAGCGUAAAUUGCUGCGCGUGCUUUUCUCACAUUGCCUCUCGCUCUGGCACGUUAA ..........(.((.(((((((((.(.((((((((((((((.....))))).))))).........(((((......)))))(((.......))))))))))))))))))).)....... ( -33.70) >DroEre_CAF1 16430 113 + 1 -------UGGGGCGUGAGCGAGAGAGCCGAUAUUUGCAUUUCGAAAAAAUGACAAAUGCUUAAUGAGCGUAAAUUGCUGCGCGUGCUUUUCUCACAUUGCCUCUCGCUCUGGCUAGUUGA -------...(((..(((((((((.((...(((((((((((.....))))).))))))........(((((......)))))(((.......)))...)))))))))))..)))...... ( -35.60) >DroYak_CAF1 27318 112 + 1 -------AGGGGCGUGAGCGAGAGGGUCGAUAUUUGCAUUUCGUAAAAAUGACAAAUGCUUAAUGAGCGUAAAUUGCUGCGCGUGCUU-UCCCACAUUGCCUCUCGCUCUGGCUCGUUUA -------..((((..((((((((((.(((.((...(((((((((....))))..))))).)).)))(((((......)))))(((...-...)))....))))))))))..))))..... ( -40.20) >consensus _______UAGGGCAUGAGCGAGAGAGUCGAUAUUUGCAUUUCGUAAAAAUGACAAAUGCCUAAUGAGCGUAAAUUGCUGCGCGUGCUUUUCUCACAUUGCCUCUCGCUCUGGCACGUUAA ...........((..(((((((((.(.((((((((((((((.....))))).))))).........(((((......)))))(((.......)))))))))))))))))..))....... (-31.48 = -31.52 + 0.04)


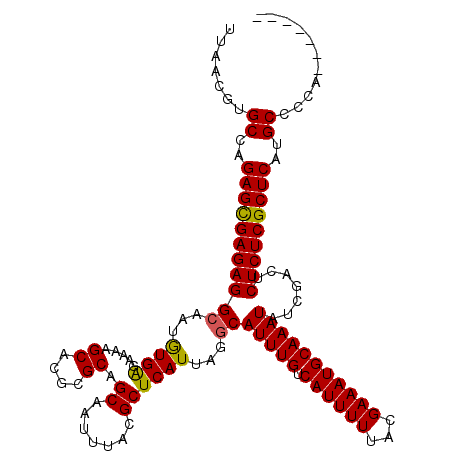
| Location | 1,994,647 – 1,994,767 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.18 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -29.32 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1994647 120 - 22224390 UUAACGUGCCAGAGUGAGAGGCAAUAUGACAAAAGCACGCGCAGCAAUUUACGCUCAUUAGGCAUUUGUCAUUUUUACGAAAUGCAAAUAUCGACUCUCUCGCUCAUGCCCUAGGAGUAC .....((((..((((((((((....((((((((.((......(((.......)))......)).)))))))).....(((.((....)).)))..))))))))))...((...)).)))) ( -31.80) >DroSec_CAF1 26691 120 - 1 UUAACGUGCUAGAGCGAGAGGCAAUGUGAGAAAAGCACGCGCAGCAAUUUACGCUCAUUAGACAUUUGUCAUUUUUACGAAAUGCAAAUAUCGACUCUCUCGCUCAUGCCCUAGGAGUAU .....(((((.(((((((((((..((((.(.....).))))..))...............((.(((((.((((((...))))))))))).))....)))))))))...((...))))))) ( -30.90) >DroSim_CAF1 20891 120 - 1 UUAACGUGCCAGAGCGAGAGGCAAUGUGAGAAAAGCACGCGCAGCAAUUUACGCUCAUUAGGCAUUUGUCAUUUUUACGAAAUGCAAAUAUCGACUCUCUCGCUCAUGCCCNNNNNNNNN .......((..(((((((((((...(((((........((...))........)))))...))(((((.((((((...))))))))))).......)))))))))..))........... ( -31.59) >DroEre_CAF1 16430 113 - 1 UCAACUAGCCAGAGCGAGAGGCAAUGUGAGAAAAGCACGCGCAGCAAUUUACGCUCAUUAAGCAUUUGUCAUUUUUUCGAAAUGCAAAUAUCGGCUCUCUCGCUCACGCCCCA------- .......((..(((((((((((..((((.......)))).))(((.......(((.....)))(((((.((((((...)))))))))))....))))))))))))..))....------- ( -31.30) >DroYak_CAF1 27318 112 - 1 UAAACGAGCCAGAGCGAGAGGCAAUGUGGGA-AAGCACGCGCAGCAAUUUACGCUCAUUAAGCAUUUGUCAUUUUUACGAAAUGCAAAUAUCGACCCUCUCGCUCACGCCCCU------- .......((..((((((((((...((((.(.-.....).)))).........(((.....)))(((((.((((((...)))))))))))......))))))))))..))....------- ( -31.90) >consensus UUAACGUGCCAGAGCGAGAGGCAAUGUGAGAAAAGCACGCGCAGCAAUUUACGCUCAUUAGGCAUUUGUCAUUUUUACGAAAUGCAAAUAUCGACUCUCUCGCUCAUGCCCCA_______ .......((..(((((((((((...((((.....((....)).((.......))))))...))(((((.((((((...))))))))))).......)))))))))..))........... (-29.32 = -29.04 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:25 2006