| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 12,259,620 – 12,259,727 |
| Length | 107 |
| Max. P | 0.801824 |

| Location | 12,259,620 – 12,259,727 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -26.80 |
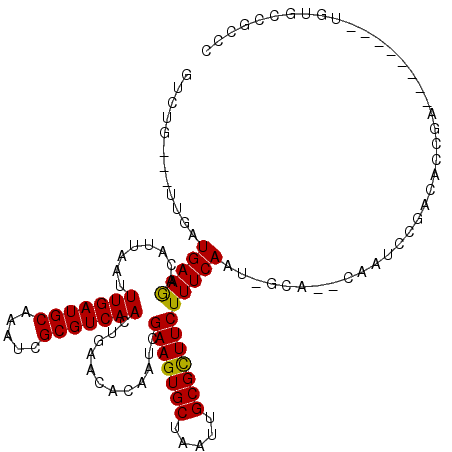
| Consensus MFE | -20.19 |
| Energy contribution | -19.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 12259620 107 + 22224390 GUCUG---UUGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAUUGGA--CAAUCCGACACCGA--------UGUGCCGCCC ((((.---.......))))......(((((((.....))))))).....((((.(((((((((.....)))))))......((((.--....))))....))--------))))...... ( -29.70) >DroPse_CAF1 17536 114 + 1 GUCUG---CUGAUGAAAACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAU-GCA--CAAUCCGACACCGAGAGACCCACGAGCCUCCA (((((---(...(((((........(((((((.....)))))))............(((((((.....))))))))))))..-)))--...((((....)).)))))............. ( -26.70) >DroGri_CAF1 8748 100 + 1 GCCCC---UUGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGUUUCUUUCAAU-GCCAGUAACCAGUAAC-AG--------U--AG----- .....---((((((....))))))..((((((.....))))))((((.........(((((((.....)))))))......(-((.........))).)-))--------)--..----- ( -17.10) >DroSec_CAF1 6216 106 + 1 GUCUG---UUGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAU-GGA--CAAUCCGACACCGA--------CGUGCCGCCC (((((---((((.............(((((((.....)))))))............(((((((.....)))))))..)))))-)))--)....((.(((...--------.))).))... ( -31.20) >DroEre_CAF1 4200 106 + 1 GUCUG---CUGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAU-GGG--CAAUCCGACACCGA--------UGUACCGCCC ((((.---.......))))......(((((((.....)))))))............(((((((.....))))))).......-(((--(......(((....--------)))...)))) ( -27.80) >DroWil_CAF1 5850 93 + 1 GUGUGCACUGGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAAAUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAU-GCAGCCAAUCA-------------------------- (..((((.((((.((........(((((((((.....)))))))))........))(((((((.....))))))).)))).)-)))..).....-------------------------- ( -28.29) >consensus GUCUG___UUGAUGAAGACAUUAAUUUGAUGCAAAUCGCGUCAACUGAACACAAUCGAAGUGCUAAUUGCGCUUCUUUCAAU_GCA__CAAUCCGACACCGA________UGUGCCGCCC ............(((((........(((((((.....)))))))............(((((((.....))))))))))))........................................ (-20.19 = -19.92 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:31:44 2006