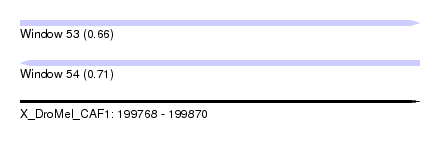
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 199,768 – 199,870 |
| Length | 102 |
| Max. P | 0.714526 |

| Location | 199,768 – 199,870 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -43.02 |
| Consensus MFE | -32.49 |
| Energy contribution | -32.46 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
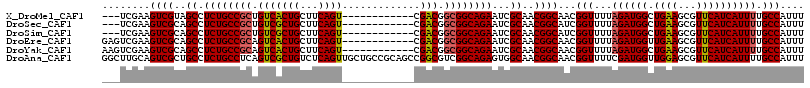
>X_DroMel_CAF1 199768 102 + 22224390 ---UCGAAGUCGUAGCCUCUGCCGCUGUCACUGCUUCAGU------------CGACGGCGGCAGAAUCGCAACGGCAACGGUUUUAGAUGGCUGAAGCGUUCAUCAUUUUGCCAUUU ---(((..(((((.((.((((((((((((((((...))))------------.))))))))))))...)).)))))..)))....(((((((.(((..........))).))))))) ( -44.40) >DroSec_CAF1 30682 102 + 1 ---UCGAAGUCGCAGCCUCUGCCGCUGUCGCUGCUUCAGU------------CGACGGCGGCAGAAUCGCAACGGCAUCGGUUUUAGAUGGCUGAAGCGUUCAUCAUUUUGCCAUUU ---((((.((((..((.((((((((((((((((...))).------------)))))))))))))...))..)))).))))....(((((((.(((..........))).))))))) ( -45.50) >DroSim_CAF1 31364 102 + 1 ---UCGAAGUCGCAGCCUCUGCCGCUGUCGCUGCUUCAGU------------CGACGGCGGCAGAAUCGCAACGGCAUCGGUUUUAGAUGGCUGAAGCGUUCAUCAUUUUGCCAUUU ---((((.((((..((.((((((((((((((((...))).------------)))))))))))))...))..)))).))))....(((((((.(((..........))).))))))) ( -45.50) >DroEre_CAF1 39255 105 + 1 GAGUCGAAGUCGCAGCCUCUGCCGCAGUCACUGCUUCAGU------------CGACGGCGGCAGAAUCGCAACGGCAACGGUUUUAGAUGGUUGAAGCGUUCAUCAUUUUGCCAUUU ((..((..((((..((.((((((((.(((((((...))))------------.))).))))))))...))..))))..))..)).(((((((.(((..........))).))))))) ( -38.20) >DroYak_CAF1 29898 105 + 1 AAGUCGAAGUCGCAGCCUCUGCCGCAGUCACUGCUUCAGU------------CGACGGCGGCAGAAUCGCAACGGCAACGGUUUUAGAUGGCUGAAGCGUUCAUCAUUUUGCCAUUU .....(((((..(((((((((((((.(((((((...))))------------.))).))))))))(((....((....))......))))))))..)).)))............... ( -40.40) >DroAna_CAF1 53325 117 + 1 GGCUUGCAGUCGCUGCCUCUGCCUCAGUCGCUGUCUCAGUUGCUGCCGCAGCCGGCGUCGGCAGAGUGGCAACGGCAACGGUUUUCGAUGGUUGGAGCGUUCAUCAUUUUGCCAUUU (((..((....)).)))((((((...((((((((..(((...)))..)))).))))...))))))(((((((((....))......(((((.........)))))...))))))).. ( -44.10) >consensus ___UCGAAGUCGCAGCCUCUGCCGCAGUCACUGCUUCAGU____________CGACGGCGGCAGAAUCGCAACGGCAACGGUUUUAGAUGGCUGAAGCGUUCAUCAUUUUGCCAUUU ........((((..((.((((((((.(((((((...)))).............))).))))))))...))..))))...(((...((((((.((((...)))))))))).))).... (-32.49 = -32.46 + -0.03)
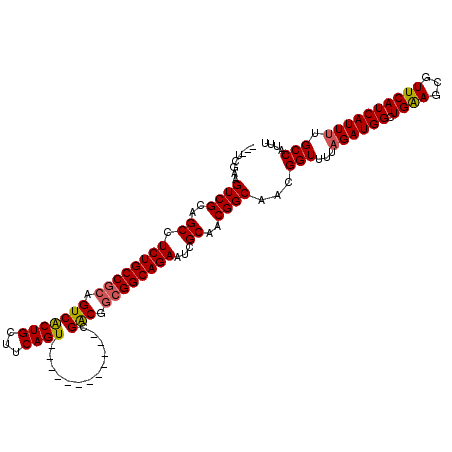
| Location | 199,768 – 199,870 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -39.99 |
| Consensus MFE | -31.03 |
| Energy contribution | -31.81 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 199768 102 - 22224390 AAAUGGCAAAAUGAUGAACGCUUCAGCCAUCUAAAACCGUUGCCGUUGCGAUUCUGCCGCCGUCG------------ACUGAAGCAGUGACAGCGGCAGAGGCUACGACUUCGA--- .((((((((...((((...((....))))))........))))))))((..(((((((((.(((.------------((((...))))))).)))))))))))..((....)).--- ( -36.50) >DroSec_CAF1 30682 102 - 1 AAAUGGCAAAAUGAUGAACGCUUCAGCCAUCUAAAACCGAUGCCGUUGCGAUUCUGCCGCCGUCG------------ACUGAAGCAGCGACAGCGGCAGAGGCUGCGACUUCGA--- ....((((....((((...((....)))))).........))))((((((.(((((((((.((((------------.(((...))))))).)))))))))..)))))).....--- ( -40.62) >DroSim_CAF1 31364 102 - 1 AAAUGGCAAAAUGAUGAACGCUUCAGCCAUCUAAAACCGAUGCCGUUGCGAUUCUGCCGCCGUCG------------ACUGAAGCAGCGACAGCGGCAGAGGCUGCGACUUCGA--- ....((((....((((...((....)))))).........))))((((((.(((((((((.((((------------.(((...))))))).)))))))))..)))))).....--- ( -40.62) >DroEre_CAF1 39255 105 - 1 AAAUGGCAAAAUGAUGAACGCUUCAACCAUCUAAAACCGUUGCCGUUGCGAUUCUGCCGCCGUCG------------ACUGAAGCAGUGACUGCGGCAGAGGCUGCGACUUCGACUC ....(((((...((((...........))))........)))))((((((.(((((((((.(((.------------((((...))))))).)))))))))..))))))........ ( -39.90) >DroYak_CAF1 29898 105 - 1 AAAUGGCAAAAUGAUGAACGCUUCAGCCAUCUAAAACCGUUGCCGUUGCGAUUCUGCCGCCGUCG------------ACUGAAGCAGUGACUGCGGCAGAGGCUGCGACUUCGACUU ..(((((....(((........))))))))........((((..((((((.(((((((((.(((.------------((((...))))))).)))))))))..))))))..)))).. ( -41.40) >DroAna_CAF1 53325 117 - 1 AAAUGGCAAAAUGAUGAACGCUCCAACCAUCGAAAACCGUUGCCGUUGCCACUCUGCCGACGCCGGCUGCGGCAGCAACUGAGACAGCGACUGAGGCAGAGGCAGCGACUGCAAGCC ....(((....(((((...........))))).....(((((((((((((.(...((((....)))).).))))))..(((...(((...)))...))).))))))).......))) ( -40.90) >consensus AAAUGGCAAAAUGAUGAACGCUUCAGCCAUCUAAAACCGUUGCCGUUGCGAUUCUGCCGCCGUCG____________ACUGAAGCAGCGACAGCGGCAGAGGCUGCGACUUCGA___ ....(((((...((((...........))))........)))))((((((.(((((((((.((((.............(((...))))))).)))))))))..))))))........ (-31.03 = -31.81 + 0.78)

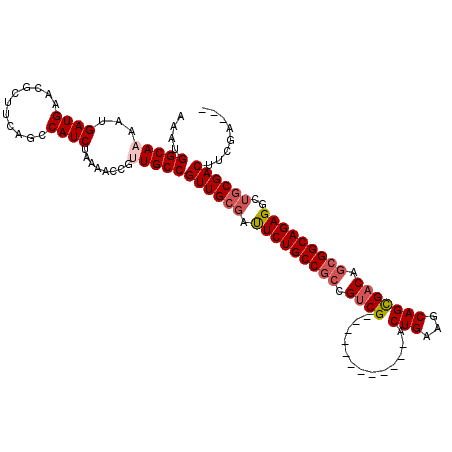
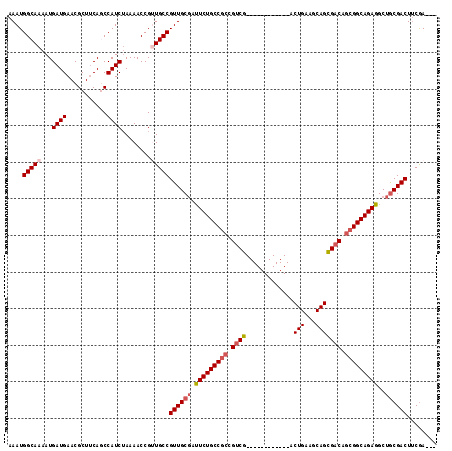
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:11 2006