| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 6,154,127 – 6,154,221 |
| Length | 94 |
| Max. P | 0.999454 |

| Location | 6,154,127 – 6,154,221 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.89 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
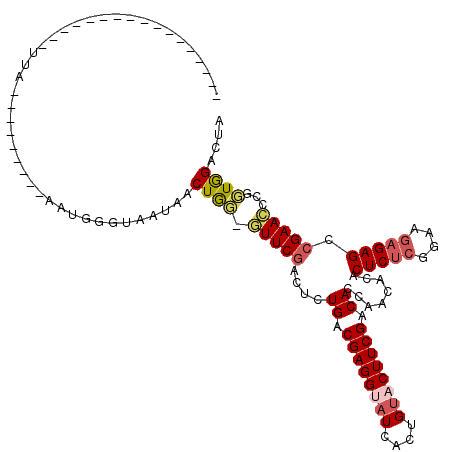
>X_DroMel_CAF1 6154127 94 + 22224390 UAGUCCAACGGAUUCGGCUCUCUUUCGAGAGUGUGUUGGUGUCGAAGUACAGUGAUACCUCGACAGAGUUGAAUACCAGUUAUGACCCAUU---------UAA----------------- ..(((.(((((((((((((((...(((((...((((((.(((......))).))))))))))).)))))))))).)).)))..))).....---------...----------------- ( -28.50) >DroSec_CAF1 7276 91 + 1 ---ACUAACGGGUUCGGCUCUCUUGCGAGAGUGUGUUGGUGUCGAAGUACAGUGAUACCUCGUCAGAGUCGAAGCCCAGUUAUUACCCAUU---------UAA----------------- ---..((((((((((((((((...(((((...((((((.(((......))).))))))))))).))))))).))))).)))).........---------...----------------- ( -31.50) >DroEre_CAF1 4992 119 + 1 UAGUCCACAGAGUUCGGCUCUCAACUGAGAGUGUGUUGGUGUCGAAGUACAGUGAUUCCUCGUCAAAGUCGAAC-GUAGAUUUUGCACUUUAAAUAUAUUUAAUUGAACUCAAUGGUUUG ....(((..(((((((((((((....))))))(((((((.((((........)))).))..(((((((((....-...))))))).))....))))).......)))))))..))).... ( -31.10) >DroYak_CAF1 4645 96 + 1 CAGUCCACCGAGUUCGGCUCUCUUCC--GAGUGUGUUGGUGUCGAAGUACAGUGAGCCCUCGCCAAAGUCGAAC-UCAGCUAUUACACAUU---------UAAUUAUU------------ .........(((((((((((((....--))).(((..(((..(........)...)))..)))...))))))))-))..............---------........------------ ( -22.50) >consensus UAGUCCAACGAGUUCGGCUCUCUUCCGAGAGUGUGUUGGUGUCGAAGUACAGUGAUACCUCGUCAAAGUCGAAC_CCAGUUAUUACACAUU_________UAA_________________ .........((((((((((((.......(((.((((((.(((......))).)))))))))...)))))))))).))........................................... (-17.83 = -18.70 + 0.87)


| Location | 6,154,127 – 6,154,221 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.89 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -15.12 |
| Energy contribution | -16.25 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 6154127 94 - 22224390 -----------------UUA---------AAUGGGUCAUAACUGGUAUUCAACUCUGUCGAGGUAUCACUGUACUUCGACACCAACACACUCUCGAAAGAGAGCCGAAUCCGUUGGACUA -----------------...---------....((((.((((.((.((((.....(((((((((((....)))))))))))........(((((....)))))..)))))))))))))). ( -35.10) >DroSec_CAF1 7276 91 - 1 -----------------UUA---------AAUGGGUAAUAACUGGGCUUCGACUCUGACGAGGUAUCACUGUACUUCGACACCAACACACUCUCGCAAGAGAGCCGAACCCGUUAGU--- -----------------...---------.........((((.(((.((((.......((((((((....))))))))...........(((((....))))).)))))))))))..--- ( -29.00) >DroEre_CAF1 4992 119 - 1 CAAACCAUUGAGUUCAAUUAAAUAUAUUUAAAGUGCAAAAUCUAC-GUUCGACUUUGACGAGGAAUCACUGUACUUCGACACCAACACACUCUCAGUUGAGAGCCGAACUCUGUGGACUA ....((((.((((((............(..(((((((...(((.(-(((.......)))).))).....)))))))..)..........(((((....)))))..)))))).)))).... ( -31.00) >DroYak_CAF1 4645 96 - 1 ------------AAUAAUUA---------AAUGUGUAAUAGCUGA-GUUCGACUUUGGCGAGGGCUCACUGUACUUCGACACCAACACACUC--GGAAGAGAGCCGAACUCGGUGGACUG ------------........---------.....((....(((((-(((((.(((((((((((((.....)).)))))...))))....(((--....))))).))))))))))..)).. ( -29.80) >consensus _________________UUA_________AAUGGGUAAUAACUGG_GUUCGACUCUGACGAGGUAUCACUGUACUUCGACACCAACACACUCUCGGAAGAGAGCCGAACCCGGUGGACUA .........................................((((.(((((....((.((((((((....)))))))).))........(((((....))))).)))))...)))).... (-15.12 = -16.25 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:20 2006