| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 543,206 – 543,355 |
| Length | 149 |
| Max. P | 0.907124 |

| Location | 543,206 – 543,322 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -42.88 |
| Consensus MFE | -31.70 |
| Energy contribution | -31.18 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 543206 116 + 22224390 GGCACUCAGAAUAGCCCGCUCAAAGUUGAGUGAAGUUGGCCAUUUCGAGUGAUUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAGGCUGUCAUGUUUGCGGUUUGAGU---- .(((..((..((((((((((((....))))))......(((....(.((((......)))).)(((((..((....)).)))))...)))..))))))..))..))).........---- ( -38.50) >DroSec_CAF1 5978 116 + 1 GCCACUCAGAAUAGCCCGCUCAAAGUUGAGUGAAGUUGGCCAUUUUGAGUGACUGUGCGCUGAUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGGCAUGUUUGCGGUUUGAGU---- ((((((((((((.(((((((((....)))))).....))).))))))))).(((((((((.....))...((....)))))))))..))).((((((.((....))))))))....---- ( -44.70) >DroSim_CAF1 6476 116 + 1 GCCACUCAGAAUAGCCCGCUCAAAGUUGAGUGAAGUUGGCCAUUUCGAGUGACUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGGCAUGUUUGCGGUUUGAGU---- ...(((((((...((.((((((....))))))......(((((((((..(.(((((((((.....))...((....))))))))).)..))))).))))......))..)))))))---- ( -43.80) >DroEre_CAF1 7669 120 + 1 GCCACUCAAAAUAGCCUGCUCAAAGUUGAGUGAAGUUGGCCGUUUUGAGUGACUGUGCGCUGGCUUCGAGAGGAAAUCGCACAGUGAGGCGAAGCUGCCAUGUUUGUGGUUUGAGUCGGG (((((((((((..(((.(((((....)))))......)))..)))))))).((((((((.(..(((....)))..).))))))))..)))((..(.(((((....)))))..)..))... ( -45.90) >DroYak_CAF1 10925 120 + 1 GCCACUCAAAAUAGCCUGCUCAAAGUUGAGUGAAGUUGCCCAUUUCGAGUGACUUUGCGCUGGAUUUGAGAGGAAACUGCACAGUGGUGCGCAGCUGCCAUGUUUGUGGCUUGAGUUGAG .((.((((((....((.((.((((((((..((((((.....))))))..)))))))).)).)).)))))).)).....((((....)))).((((((((((....)))))...))))).. ( -41.50) >consensus GCCACUCAGAAUAGCCCGCUCAAAGUUGAGUGAAGUUGGCCAUUUCGAGUGACUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGCCAUGUUUGCGGUUUGAGU____ (((((((.((((.(((((((((....)))))).....))).)))).)))))(((((((.((.......))((....)))))))))...)).((((((.((....))))))))........ (-31.70 = -31.18 + -0.52)

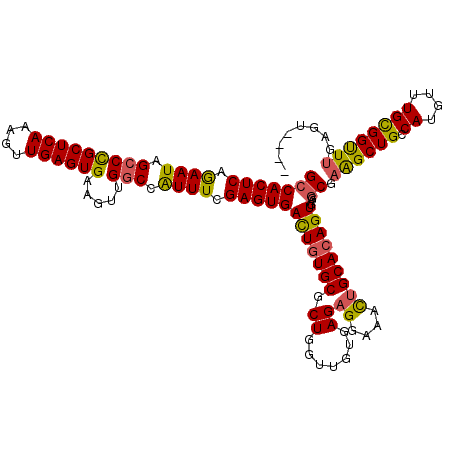
| Location | 543,206 – 543,322 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -23.49 |
| Energy contribution | -24.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 543206 116 - 22224390 ----ACUCAAACCGCAAACAUGACAGCCUCGCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAAUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGCC ----........(((.....(((..((...)).))).((((.((......))))))....)))......((((((.(((((((..((.((........))))..))))))).)))))).. ( -25.80) >DroSec_CAF1 5978 116 - 1 ----ACUCAAACCGCAAACAUGCCAGCUUCGCCUCACUGUGCAGUUUCCUCUCACAAUCAGCGCACAGUCACUCAAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGC ----.........(((....))).......(((..(((((((.(((.............)))))))))).(((((.(((((((..((.((........))))..))))))).)))))))) ( -31.42) >DroSim_CAF1 6476 116 - 1 ----ACUCAAACCGCAAACAUGCCAGCUUCGCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAGUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGC ----.........(((....))).......(((..(((((((.(((.............)))))))))).(((((.(((((((..((.((........))))..))))))).)))))))) ( -30.72) >DroEre_CAF1 7669 120 - 1 CCCGACUCAAACCACAAACAUGGCAGCUUCGCCUCACUGUGCGAUUUCCUCUCGAAGCCAGCGCACAGUCACUCAAAACGGCCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGC .....................(((......)))..((((((((.((((.....))))....))))))))(((((((((..(((...((((........))))..)))..))))))))).. ( -36.00) >DroYak_CAF1 10925 120 - 1 CUCAACUCAAGCCACAAACAUGGCAGCUGCGCACCACUGUGCAGUUUCCUCUCAAAUCCAGCGCAAAGUCACUCGAAAUGGGCAACUUCACUCAACUUUGAGCAGGCUAUUUUGAGUGGC ..........((((......))))(((((((((....))))))))).....................((((((((((((((.(...((((........))))..).)))))))))))))) ( -37.10) >consensus ____ACUCAAACCGCAAACAUGCCAGCUUCGCCUCACUGUGCAGUUUCCUCUCACAACCAGCGCACAGUCACUCGAAAUGGCCAACUUCACUCAACUUUGAGCGGGCUAUUCUGAGUGGC ...................................(((((((....................)))))))((((((.(((((((..((.((........))))..))))))).)))))).. (-23.49 = -24.05 + 0.56)

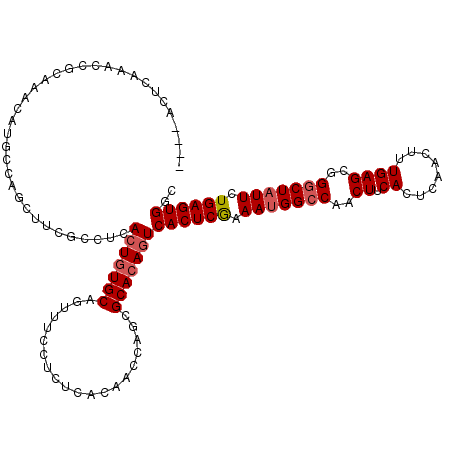
| Location | 543,246 – 543,355 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -26.37 |
| Energy contribution | -25.93 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 543246 109 + 22224390 CAUUUCGAGUGAUUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAGGCUGUCAUGUUUGCGGUUUGAGU-------CCAUGGCAUUGUGAGUUGGAGCCGAAAAUAUGU ..(((((..(.(((((((((.....))...((....))))))))).)..)))))....(((((((.((((((.((.-------.(((......)))..)).)))))).))))))). ( -34.90) >DroSec_CAF1 6018 109 + 1 CAUUUUGAGUGACUGUGCGCUGAUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGGCAUGUUUGCGGUUUGAGU-------CCGUGGCAUGGUGAGUUGGAGCCGGAAAUAUGU (((((((..(.(((((((((.....))...((....))))))))).)..))))).))((((((((.((((((.((.-------.((........))..)).)))))).)))))))) ( -37.40) >DroSim_CAF1 6516 109 + 1 CAUUUCGAGUGACUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGGCAUGUUUGCGGUUUGAGU-------CCGUGGCAUGGUGAGUUGGAGCCGGAAAUAUGU (((((((..(.(((((((((.....))...((....))))))))).)..))))).))((((((((.((((((.((.-------.((........))..)).)))))).)))))))) ( -39.50) >DroEre_CAF1 7709 116 + 1 CGUUUUGAGUGACUGUGCGCUGGCUUCGAGAGGAAAUCGCACAGUGAGGCGAAGCUGCCAUGUUUGUGGUUUGAGUCGGGCUGCCACGGCAUGUUGAGUUGAUGCCGAAAACAUGU .((((((..(.((((((((.(..(((....)))..).)))))))).)..))))))...(((((((.(((((..(.((((..(((....)))..)))).)..).)))).))))))). ( -43.10) >DroYak_CAF1 10965 116 + 1 CAUUUCGAGUGACUUUGCGCUGGAUUUGAGAGGAAACUGCACAGUGGUGCGCAGCUGCCAUGUUUGUGGCUUGAGUUGAGCUGCCAAGGCAUGUUGAGUUGCAGCCGAUAACAUGU ((..((.((((......)))).))..))..((....))((((....))))((((((..((((((((..((((.....))))..)..)))))))...)))))).............. ( -37.20) >consensus CAUUUCGAGUGACUGUGCGCUGGUUGUGAGAGGAAACUGCACAGUGAGGCGAAGCUGCCAUGUUUGCGGUUUGAGU_______CCAUGGCAUGGUGAGUUGGAGCCGAAAAUAUGU ..(((((..(.(((((((.((.......))((....))))))))).)..)))))....(((((((.((((((.((........(....).........)).)))))).))))))). (-26.37 = -25.93 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:21 2006