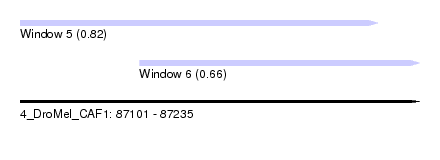
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 87,101 – 87,235 |
| Length | 134 |
| Max. P | 0.821410 |

| Location | 87,101 – 87,221 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.47 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -31.80 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 87101 120 + 1281640 UUUCGAUAUCUUUCGCAAUCGAGUCCAAGGCCAGCUUGUUAACAAGCUGCUUUAGAUCGGCACCACUAACUUCAUUAGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGGACUUGCG .((((((..........))))))...(((((.((((((....))))))))))).((((((((..(((((.....))))).....))))))))..(((((..(((.....)))..))))). ( -37.70) >DroSec_CAF1 29400 120 + 1 UUUCGAUAUCUUUUGCAAUCGAGUCCAAGGCCAGUUUGUUAACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGGACUUGUG .((((((..........))))))...(((((.((((((....))))))))))).((((((((..((((((...)))))).....))))))))..(((((..(((.....)))..))))). ( -34.80) >DroEre_CAF1 46617 120 + 1 UUUCAAUAUCUUUUGCAAUCGAGUCCAAGGCCAGUUUGUUAACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUUGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUCCGAACUUGCG ..((.((((...((((((.((((((.(((((.((((((....))))))))))).))).(....).......))).))))))..)))).))....(((((..((.......))..))))). ( -26.70) >DroYak_CAF1 52792 120 + 1 UUUCAAUGUCUUUCGCAAUUGAGUCCAGAGCAAGUUUGUUAACUAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGAACUUGCG .((((((((.....)).))))))......(((((((((.........(((((..((((((((..((((((...)))))).....))))))))....))))).........))))))))). ( -32.37) >consensus UUUCAAUAUCUUUCGCAAUCGAGUCCAAGGCCAGUUUGUUAACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGAACUUGCG .((((((..........))))))...(((((.((((((....))))))))))).((((((((..((((((...)))))).....))))))))..(((((..(((.....)))..))))). (-31.80 = -31.93 + 0.13)

| Location | 87,141 – 87,235 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.57 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -23.96 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 87141 94 + 1281640 AACAAGCUGCUUUAGAUCGGCACCACUAACUUCAUUAGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGGACUUGCGAUUGCUGAGCAUAA .......(((((..((((((((..(((((.....))))).....)))))))).((((((..(((.....)))..))))))......)))))... ( -26.40) >DroSec_CAF1 29440 94 + 1 AACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGGACUUGUGAUUGCUGAGCAUAA .......(((((..((((((((..((((((...)))))).....))))))))..((((.(((((.((.....)).))))).)))).)))))... ( -27.70) >DroEre_CAF1 46657 94 + 1 AACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUUGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUCCGAACUUGCGAUUGUUGAGCAUAA .......(((((..((((((((......((.......)).....)))))))).((((((..((.......))..))))))......)))))... ( -20.80) >DroYak_CAF1 52832 94 + 1 AACUAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGAACUUGCGAUUGUUGAGCAUAA .......(((((..((((((((..((((((...)))))).....)))))))).((((((..(((.....)))..))))))......)))))... ( -26.10) >consensus AACAAGCUGCUUUAGAUCGGCACCACUAACUUCGUUGGUAAUAAUGUCGGUCAUGCGAGCACGAAUCUUUCGAACUUGCGAUUGCUGAGCAUAA .......(((((..((((((((..((((((...)))))).....)))))))).((((((..(((.....)))..))))))......)))))... (-23.96 = -24.40 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:50 2006