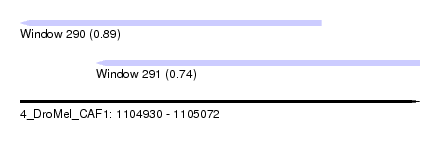
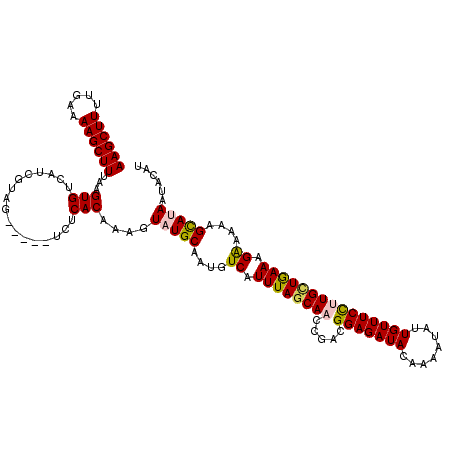
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,104,930 – 1,105,072 |
| Length | 142 |
| Max. P | 0.894810 |

| Location | 1,104,930 – 1,105,037 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.60 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -18.25 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1104930 107 - 1281640 CACAAAGUAUGCAAUGUCAUUUAGCAACCAACGGAGAUACAAAAUAUUGUUUCCGUGCUAAAAGGAAAAGCAUAAUACAUUUAUAAAUCGUGAAAAUGUGAUA-------------GUGU (((....(((((....((.((((((.....(((((((((........))))))))))))))).))....))))).(((((((.((.....)).)))))))...-------------))). ( -25.20) >DroSec_CAF1 18438 107 - 1 CACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAAAUAUUGUUUCCUUGUUGAAAGAAAAAGUAUAAUACAUUUAUAAAUCGUGAAGAUGUGAUA-------------GUGU (((....(((((....((.((((((((.....(((((((........))))))))))))))).))....))))).(((((((.((.....)).)))))))...-------------))). ( -20.30) >DroEre_CAF1 18646 120 - 1 CACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAUAUAUUGUUUCUAUGCUGAAAGAAAAAGCAAAAAACGUGUACAAAACGUGAAAAUGUGAUAUUUCUUACAGAGAGUGU ((((.....(((....((.(((((((.((...))(((.((((....)))).))).))))))).))....)))....((((.......)))).....))))...((((.....)))).... ( -22.30) >DroYak_CAF1 9746 120 - 1 CACAAAGUAUGCAAUGUCAUUUAACAACCGACGGAGAUAUUAUAUAUUGUUUCCUUGGUGAAAGAAAAAGCAGAAUACAAUUAUAUAACGUGAAAAUGUGAUACUUUUUACAGAAAGUGU ......((((....(((..(((....(((((.(((((((........)))))))))))).....)))..)))..)))).........((((....)))).((((((((....)))))))) ( -25.80) >consensus CACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAAAUAUUGUUUCCUUGCUGAAAGAAAAAGCAUAAUACAUUUAUAAAACGUGAAAAUGUGAUA_____________GUGU (((....(((((....((.((((((((.....(((((((........))))))))))))))).))....))))).(((((((.((.....)).)))))))................))). (-18.25 = -18.88 + 0.62)

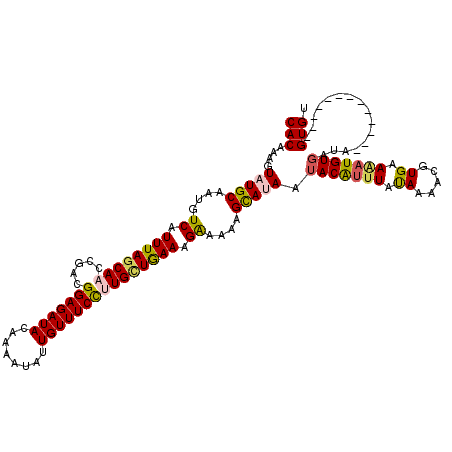
| Location | 1,104,957 – 1,105,072 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1104957 115 - 1281640 AAGCUUUUGAAAAGCUUAAAGUGUCAUCUUAG-----UUUCACAAAGUAUGCAAUGUCAUUUAGCAACCAACGGAGAUACAAAAUAUUGUUUCCGUGCUAAAAGGAAAAGCAUAAUACAU ((((((.....))))))...(((..((....)-----)..)))....(((((....((.((((((.....(((((((((........))))))))))))))).))....)))))...... ( -25.60) >DroSec_CAF1 18465 115 - 1 AAGCUUUUAAAAAGCUUUAAGUGUCAUCUUAG-----UUUCACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAAAUAUUGUUUCCUUGUUGAAAGAAAAAGUAUAAUACAU ((((((.....))))))...(((..((....)-----)..)))....(((((....((.((((((((.....(((((((........))))))))))))))).))....)))))...... ( -20.00) >DroEre_CAF1 18686 115 - 1 AAGCUUUUGAAAAGCUUUGAGUGUCACCGUAG-----UCUCACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAUAUAUUGUUUCUAUGCUGAAAGAAAAAGCAAAAAACGU ..((((((.....((((((.(.(.(......)-----.).).))))))........((.(((((((.((...))(((.((((....)))).))).))))))).))))))))......... ( -23.00) >DroYak_CAF1 9786 120 - 1 AAGCUUUUGAAAAGCUUUAAGUGUCAUCGUAGUAUUGUCUCACAAAGUAUGCAAUGUCAUUUAACAACCGACGGAGAUAUUAUAUAUUGUUUCCUUGGUGAAAGAAAAAGCAGAAUACAA ..((((((........(((((((.(((.(((...(((.....)))....))).))).)))))))..(((((.(((((((........))))))))))))......))))))......... ( -28.50) >consensus AAGCUUUUGAAAAGCUUUAAGUGUCAUCGUAG_____UCUCACAAAGUAUGCAAUGUCAUUUAGCAACCGACGGAGAUACAAAAUAUUGUUUCCUUGCUGAAAGAAAAAGCAUAAUACAU ((((((.....))))))...(((.................)))....(((((....((.((((((((.....(((((((........))))))))))))))).))....)))))...... (-19.13 = -19.50 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:26 2006