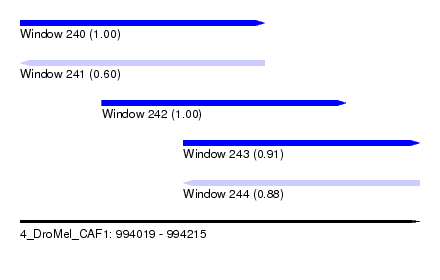
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 994,019 – 994,215 |
| Length | 196 |
| Max. P | 0.998841 |

| Location | 994,019 – 994,139 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
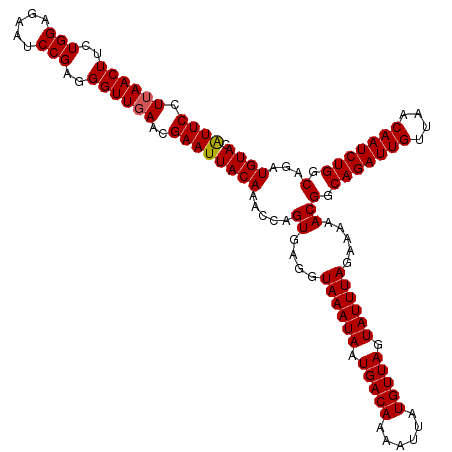
>4_DroMel_CAF1 994019 120 + 1281640 AUUCCUUAACUUCUGGAGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUAC ((((.((((((..(((.....)))..))))))..))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). ( -29.00) >DroSec_CAF1 33082 120 + 1 AUUCGUUAACUUCUGGAGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUAC (((((((((((..(((.....)))..))))).))))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). ( -32.80) >DroSim_CAF1 27792 120 + 1 GUUCGUUAACUUCUGGAGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUAC (((((((((((..(((.....)))..))))).))))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). ( -33.10) >DroEre_CAF1 31520 120 + 1 AUUCCUUAACUUCUGGGGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGCUAACAAUCUGGCAGAUGUAC ((((.((((((..(((.....)))..))))))..))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). ( -27.30) >DroYak_CAF1 29852 120 + 1 AUUCCUAAACUUCUGGGGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUAC ((((...((((..(((.....)))..))))....))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). ( -26.50) >consensus AUUCCUUAACUUCUGGAGAAUCCGAGGGUUGAACGAAUUACAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUAC ((((.((((((..(((.....)))..))))))..))))((((.....((....((((((.(((((......))))).)))))).....))(.(((((((....))))))).)...)))). (-27.32 = -27.36 + 0.04)

| Location | 994,019 – 994,139 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -21.38 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 994019 120 - 1281640 GUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCUCCAGAAGUUAAGGAAU ..........(((((((....)))))))((.......((((((.(.(((......))).).)))))).....((((...(.((((((.........))))))).)))).......))... ( -20.50) >DroSec_CAF1 33082 120 - 1 GUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCUCCAGAAGUUAACGAAU .((((...(((((((((....))))))..........((((((.(.(((......))).).)))))).......))).))))(((((((.(((..((((.....)).)).)))))))))) ( -22.50) >DroSim_CAF1 27792 120 - 1 GUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCUCCAGAAGUUAACGAAC .((((...(((((((((....))))))..........((((((.(.(((......))).).)))))).......))).)))).((((((.(((..((((.....)).)).))))))))). ( -21.90) >DroEre_CAF1 31520 120 - 1 GUACAUCUGCCAGAUUGUUAGCAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCCCCAGAAGUUAAGGAAU ..((.((((.(((((((....)))))))(((...............(((......))).(((((..(((.....)))..)))))............)))......)))).))........ ( -20.00) >DroYak_CAF1 29852 120 - 1 GUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCCCCAGAAGUUUAGGAAU ........(.(((((((....))))))).)...(((((((((....(((......)))..............((((...(.((((((.........))))))).))))..))))))))). ( -22.00) >consensus GUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAAAUACUAACAUAAUUUUGUCAUUAUUUACCUCACUGGUUUGUAAUUCGUUCAACCCUCGGAUUCUCCAGAAGUUAAGGAAU ..((.((((.(((((((....)))))))(((...............(((......))).(((((..(((.....)))..)))))............)))......)))).))........ (-19.76 = -19.76 + -0.00)

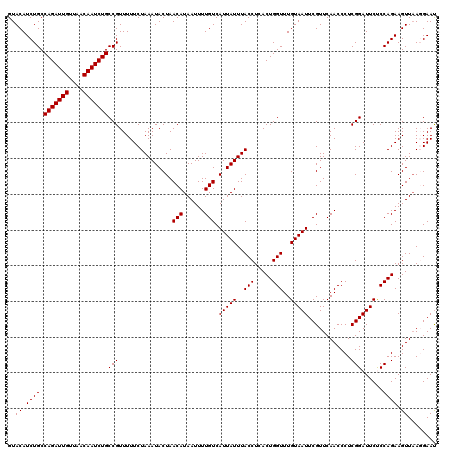
| Location | 994,059 – 994,179 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.46 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
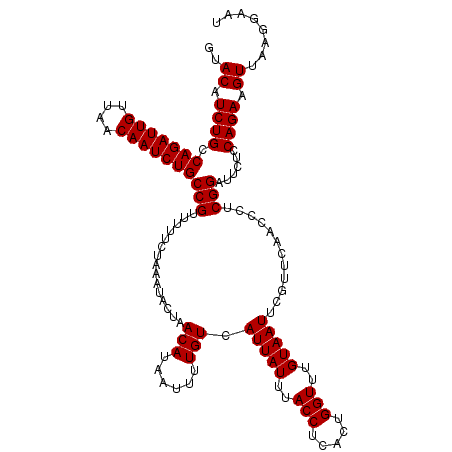
>4_DroMel_CAF1 994059 120 + 1281640 CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUACAAAUCGAAUAAGCAGGGACGAGUGC ....((..((((.((((((.(((((......))))).)))))).......(.(((((((....))))))).).......))))....((((((....))))))......))......... ( -28.00) >DroSec_CAF1 33122 120 + 1 CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGC ....((..((((.((((((.(((((......))))).)))))).......(.(((((((....))))))).).......))))....((((((....))))))......))......... ( -27.70) >DroSim_CAF1 27832 120 + 1 CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGC ....((..((((.((((((.(((((......))))).)))))).......(.(((((((....))))))).).......))))....((((((....))))))......))......... ( -27.70) >DroEre_CAF1 31560 120 + 1 CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGCUAACAAUCUGGCAGAUGUACUCAAACUUCCGAUCAAAAUCGAGUAAGCAGGGACGAGUGC ....((..((((.((((((.(((((......))))).)))))).......(.(((((((....))))))).).......))))..((((((((....)))).).)))..))......... ( -25.30) >DroYak_CAF1 29892 120 + 1 CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUUGAUCAAAAUCGAGUAAGCAGGGACGAGUGC ...(((.....)))(((((.(((((......))))).)))))........(.(((((((....))))))).)....((((((......(((((....)))))((........)))))))) ( -25.40) >consensus CAAACCAGUGAGGUAAAUAAUGACAAAAUUAUGUUAGUAUUUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGC ....((..((((.((((((.(((((......))))).)))))).......(.(((((((....))))))).).......))))....((((((....))))))......))......... (-26.66 = -26.46 + -0.20)

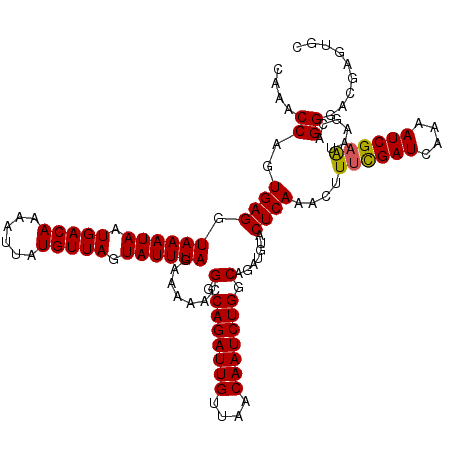
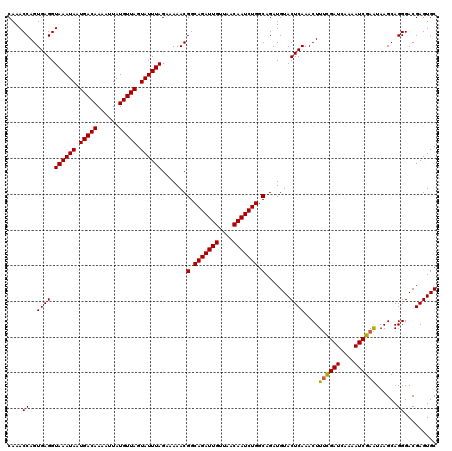
| Location | 994,099 – 994,215 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.76 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 994099 116 + 1281640 UUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUACAAAUCGAAUAAGCAGGGACGAGUGCCUGAGGUUUAACGCACU----GCGGGGUUGUUUGUGAGAG ..........(.(((((((....))))))).).......((((....((((((....))))))(((((((...(((((((.(........).)))))----.))...))))))))))).. ( -31.60) >DroSec_CAF1 33162 116 + 1 UUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGCCUGAGGUUUAACGCACU----GCGGGGUUGUUUGUAAGAG ................(((..((((((((.((((.(((....(((((((((((....))))).....((((.(....).))))))))))...)))))----)).))))))))..)))... ( -31.00) >DroSim_CAF1 27872 116 + 1 UUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGCCUGAGGUUUAACGCACU----GCGGGGUUGUUUGUAAGAG ................(((..((((((((.((((.(((....(((((((((((....))))).....((((.(....).))))))))))...)))))----)).))))))))..)))... ( -31.00) >DroEre_CAF1 31600 120 + 1 UUAGAAAAACGGCAGAUUGCUAACAAUCUGGCAGAUGUACUCAAACUUCCGAUCAAAAUCGAGUAAGCAGGGACGAGUGCCUGAGGUUUAACGCAUUAAAGGCGGGGUUGUUUGCAAGAG ............(((((((....)))))))(((((((.(((((((((((((((....)))).......(((.(....).))))))))))..(((.......))))))))))))))..... ( -31.90) >DroYak_CAF1 29932 120 + 1 UUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUUGAUCAAAAUCGAGUAAGCAGGGACGAGUGCCUGAGGUUUAACGCAUCAAAGGCGGGGUUGUUUGUAAGAG ............(((((((....)))))))(((((((.((((...(((((((((.(((((.......((((.(....).)))).)))))...).))))))))..)))))))))))..... ( -30.10) >consensus UUAGAAAAACGGCAGAUUGUUAACAAUCUGGCAGAUGUACUCAAACUUUCGAUCAAAAUCGAAUAAGCAGGGACGAGUGCCUGAGGUUUAACGCACU____GCGGGGUUGUUUGUAAGAG ............(((((((....)))))))(((((((.(((((((((((((((....))))))....((((.(....).)))).)))))..(((.......))))))))))))))..... (-31.12 = -30.76 + -0.36)

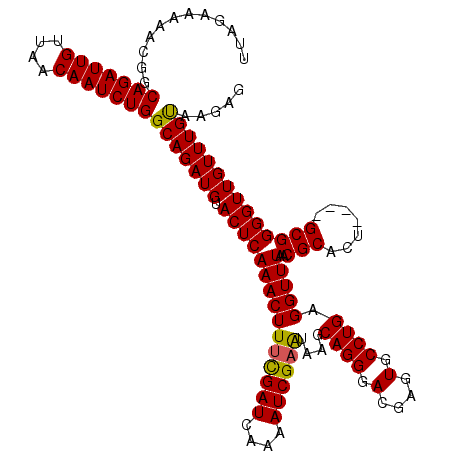
| Location | 994,099 – 994,215 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 994099 116 - 1281640 CUCUCACAAACAACCCCGC----AGUGCGUUAAACCUCAGGCACUCGUCCCUGCUUAUUCGAUUUGUAUCGAAAGUUUGAGUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAA .......((((......((----(((((.((((((...(((((........))))).((((((....)))))).))))))))))...)))(((((((....)))))))..))))...... ( -26.90) >DroSec_CAF1 33162 116 - 1 CUCUUACAAACAACCCCGC----AGUGCGUUAAACCUCAGGCACUCGUCCCUGCUUAUUCGAUUUUGAUCGAAAGUUUGAGUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAA .......((((......((----(((((.((((((...(((((........))))).((((((....)))))).))))))))))...)))(((((((....)))))))..))))...... ( -26.60) >DroSim_CAF1 27872 116 - 1 CUCUUACAAACAACCCCGC----AGUGCGUUAAACCUCAGGCACUCGUCCCUGCUUAUUCGAUUUUGAUCGAAAGUUUGAGUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAA .......((((......((----(((((.((((((...(((((........))))).((((((....)))))).))))))))))...)))(((((((....)))))))..))))...... ( -26.60) >DroEre_CAF1 31600 120 - 1 CUCUUGCAAACAACCCCGCCUUUAAUGCGUUAAACCUCAGGCACUCGUCCCUGCUUACUCGAUUUUGAUCGGAAGUUUGAGUACAUCUGCCAGAUUGUUAGCAAUCUGCCGUUUUUCUAA .......((((.....(((.......)))..........((((((((.....((((.(.((((....))))))))).))))).........((((((....)))))))))))))...... ( -23.30) >DroYak_CAF1 29932 120 - 1 CUCUUACAAACAACCCCGCCUUUGAUGCGUUAAACCUCAGGCACUCGUCCCUGCUUACUCGAUUUUGAUCAAAAGUUUGAGUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAA .......((((......((....((((..........((((........))))..(((((((((((.....)))).))))))))))).))(((((((....)))))))..))))...... ( -22.60) >consensus CUCUUACAAACAACCCCGC____AGUGCGUUAAACCUCAGGCACUCGUCCCUGCUUAUUCGAUUUUGAUCGAAAGUUUGAGUACAUCUGCCAGAUUGUUAACAAUCUGCCGUUUUUCUAA .......((((.....(((.......)))..........((((((((.....((((..(((((....))))))))).))))).........((((((....)))))))))))))...... (-21.96 = -22.00 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:20 2006