
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 985,635 – 985,890 |
| Length | 255 |
| Max. P | 0.966873 |

| Location | 985,635 – 985,755 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 985635 120 + 1281640 CGCUCCGCUUCCCUGCGGGUAAUCAUGCCACAUCGUCGGGAACUCACUGGCAUGGCGCCAGCUCUUCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACAAGUAAUGCCAAGAGCA ((((((((......)))))....(((((((....((.....))....))))))))))...((((((..((.(((((((........))((((((...))))))))))).))..)))))). ( -36.90) >DroSec_CAF1 18618 120 + 1 CGCUCCGCUUCCCUGCGGGUAAUCAUGCCACAUCGUCGGGAACUCACUGGCAUGGCGCCCGCUCUCCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACCAGUAAUGCCAAGAGCG (((((.((......((((((..((((((((....((.....))....)))))))).)))))).................(((((...(((((((...))))))).)))))))...))))) ( -41.30) >DroSim_CAF1 16262 120 + 1 CGCUCCGCUUCCCUGCGGGUAAUCAUGCCACAUCGUCGGGAACUCACUGGCAUGGCGCCCGCUCUCCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACAAGUAAUGCCAAGAGCG (((((.((......((((((..((((((((....((.....))....)))))))).)))))).................(((((....((((((...))))))..)))))))...))))) ( -37.70) >DroEre_CAF1 19462 120 + 1 CGCUCCGCUUCCCUUCGGGUUAUCAUGCCACAUCGUCGGGAACUCACUGGCAUGGCGCCAGCUCUUCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACAAGUAAUGCCCAGAGCA .((((.(((((((..(((((......)))....))..)))))....(((((.....))))).....................))..((((((((.(((....))))).)))))).)))). ( -35.10) >DroYak_CAF1 19807 120 + 1 CGCUCCGCUUCUCUGCGGGUUAUCAUGCCACAUCGUCGUGAACUUACUGGCAUUGCGCCAGCUCUUCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACAAGUAAUGCCAAGAGCA .(((((((......))).....(((((.(.....).)))))......(((((((((.((.((....................))..))((((((...))))))..))))))))).)))). ( -34.15) >consensus CGCUCCGCUUCCCUGCGGGUAAUCAUGCCACAUCGUCGGGAACUCACUGGCAUGGCGCCAGCUCUUCUCAAUAUUUCCAAUUGCACGGGUGUACAUUGUACACAAGUAAUGCCAAGAGCA ((((((((......)))))....(((((((....((.....))....))))))))))...(((((...((.(((((((........))((((((...))))))))))).))...))))). (-32.32 = -32.92 + 0.60)

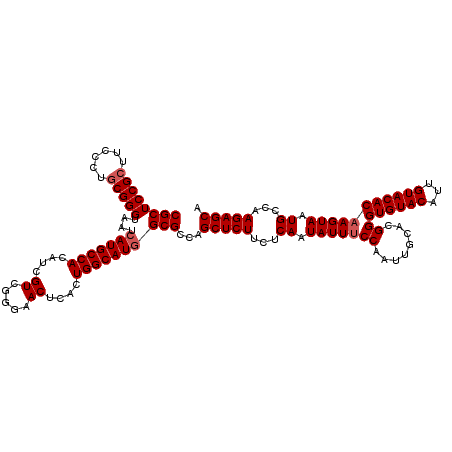
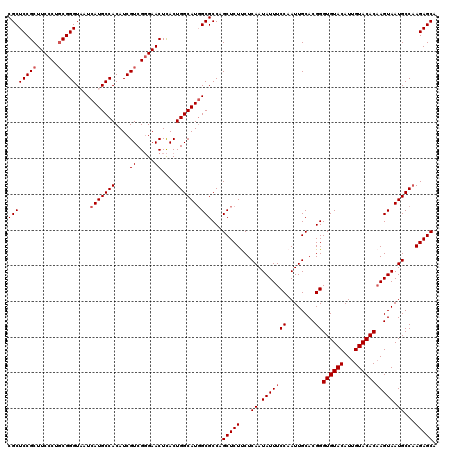
| Location | 985,715 – 985,835 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -25.86 |
| Energy contribution | -26.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 985715 120 - 1281640 CAAGAGCGCCUUUGCUUGGCGCAAAUUUCAAAUACAUUACUAAAACAAUUCAGUUACAACGAGAUACACAACAGGAGAGUUGCUCUUGGCAUUACUUGUGUACAAUGUACACCCGUGCAA ((((((((.((((.((((......(((((...((......)).(((......))).....)))))......)))))))).))))))))((((.....((((((...))))))..)))).. ( -27.40) >DroSec_CAF1 18698 120 - 1 CAGGAGCGCCUGUGCUGGGGGCAAAUAUCAGAUUCUUUUCUAAAGCAAUUCAGUGUCAACGAGAUACACAACAGGAGAGUCGCUCUUGGCAUUACUGGUGUACAAUGUACACCCGUGCAA ((((((((.((.(.(((..(.........(((......)))...........(((((.....))))).)..))).).)).))))))))((((....(((((((...))))))).)))).. ( -40.20) >DroSim_CAF1 16342 120 - 1 CAAGAGCGCCUUUGCUUGGCGCAAAUAUCAAAUACAUUACUAAAACAAUUCAGUUACAACGAGAUACACAACAGGAGAGUCGCUCUUGGCAUUACUUGUGUACAAUGUACACCCGUGCAA ((((((((.((((.((((.......((((.........(((.((....)).)))........)))).....)))))))).))))))))((((.....((((((...))))))..)))).. ( -28.33) >DroEre_CAF1 19542 120 - 1 CAAGAGCGCCUUUGCUUGGCGCAAAUCUCAAAUACAUUACUGAAACAAUUUAGUUAUAACGAGAUACAUAACAGGAGAGUUGCUCUGGGCAUUACUUGUGUACAAUGUACACCCGUGCAA .....((((...((((..(.((((.((((.........((((((....))))))............(......))))).)))).)..))))......((((((...))))))..)))).. ( -30.80) >DroYak_CAF1 19887 120 - 1 CAAGAGCGCCUUUGUUUGGCGCAAAUCUCAAAUACAUUAUUAAAACAAUUCAGUUAUAACGAGAUACACAACAGGAGAGUUGCUCUUGGCAUUACUUGUGUACAAUGUACACCCGUGCAA .....(((((.......)))))..(((((.((((...))))..(((......))).....)))))......((((((.....))))))((((.....((((((...))))))..)))).. ( -29.00) >consensus CAAGAGCGCCUUUGCUUGGCGCAAAUAUCAAAUACAUUACUAAAACAAUUCAGUUACAACGAGAUACACAACAGGAGAGUUGCUCUUGGCAUUACUUGUGUACAAUGUACACCCGUGCAA ((((((((.((((.((((......(((((...............................)))))......)))))))).))))))))((((.....((((((...))))))..)))).. (-25.86 = -26.22 + 0.36)

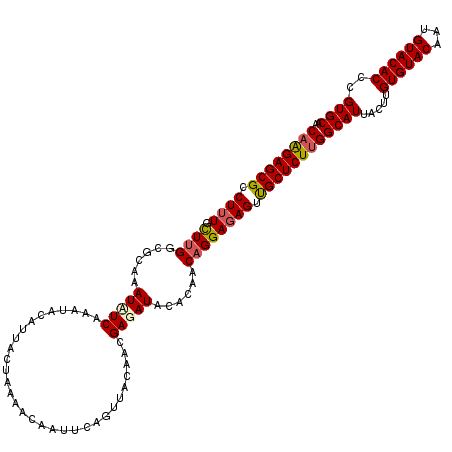
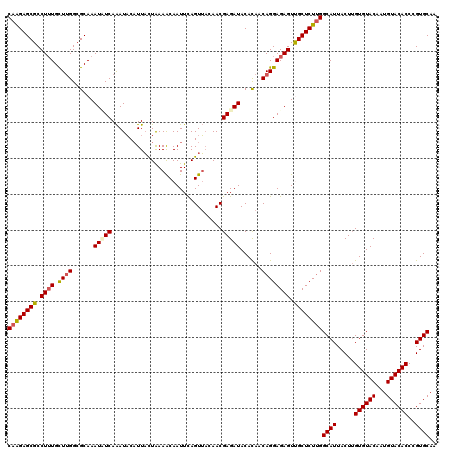
| Location | 985,795 – 985,890 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.02 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -24.81 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 985795 95 + 1281640 GUAAUGUAUUUGAAAUUUGCGCCAAGCAAAGGCGCUCUUGACCUUCAAUGUGCAGGGAUACAAUCUGCACUCCGUACAGCAAAACGGAGCUUACC ((((.((..(((((.((.(((((.......)))))....))..))))).(((((((.......)))))))(((((........))))))))))). ( -26.90) >DroSim_CAF1 16422 95 + 1 GUAAUGUAUUUGAUAUUUGCGCCAAGCAAAGGCGCUCUUGACCUUCAAUGUGCAAGGAUACAAUCUGCACUCCGUACAGCAAAACGGAGCUAACC (((.((((((........(((((.......))))).((((..(......)..))))))))))...))).((((((........))))))...... ( -25.20) >DroEre_CAF1 19622 95 + 1 GUAAUGUAUUUGAGAUUUGCGCCAAGCAAAGGCGCUCUUGACCUUCAAUGUGCAAGGAUACAAUCUGCACUCCGUACAGCAAAACGGAGCUAACC (((.((((((..(((...(((((.......))))))))..)(((((.....).)))))))))...))).((((((........))))))...... ( -26.10) >DroYak_CAF1 19967 95 + 1 AUAAUGUAUUUGAGAUUUGCGCCAAACAAAGGCGCUCUUGACCUUCAAUGUGCAAGGAUACAAUCUGCACUCCGUACAGCAAAACGGAGCUAACC ....((((((..(((...(((((.......))))))))..)(((((.....).))))))))).......((((((........))))))...... ( -25.60) >consensus GUAAUGUAUUUGAGAUUUGCGCCAAGCAAAGGCGCUCUUGACCUUCAAUGUGCAAGGAUACAAUCUGCACUCCGUACAGCAAAACGGAGCUAACC ....((((((........(((((.......))))).((((..(......)..)))))))))).......((((((........))))))...... (-24.81 = -24.62 + -0.19)

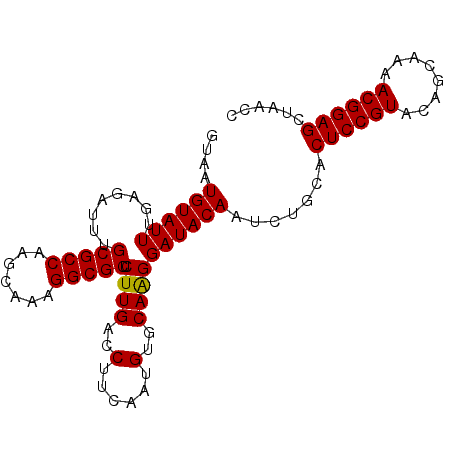
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:11 2006