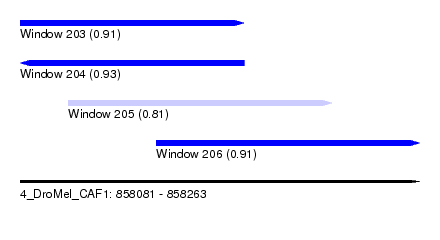
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 858,081 – 858,263 |
| Length | 182 |
| Max. P | 0.925160 |

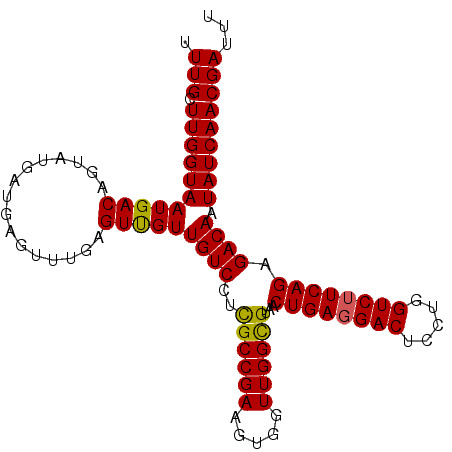
| Location | 858,081 – 858,183 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 96.27 |
| Mean single sequence MFE | -29.55 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.95 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 858081 102 + 1281640 UUUGCUUGGUAAUGACAGUAUGAUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGACGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUU .(((.((((((.((......(((.((.(..((((((((((...((((((.....))))))..)).))))))))..).)).)))....)).)))))))))... ( -27.80) >DroSec_CAF1 38304 102 + 1 UUUGCUUGGUAAUGACAGUAUGAUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUU .(((.((((((..(((((..(((.....)))..)))))(((..((((((.....))))))...((((((((.....)))))))).)))..)))))))))... ( -28.00) >DroSim_CAF1 33399 102 + 1 UUUGCUUGGUAAUGACAGUAUGAUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUU .(((.((((((..(((((..(((.....)))..)))))(((..((((((.....))))))...((((((((.....)))))))).)))..)))))))))... ( -28.00) >DroEre_CAF1 38116 102 + 1 UUUGUUUGGUAAUGACUGUAUGAUGAGUUUGAGUCGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUU .((((((((((((((((..............)))))))).)).((((((.....))))))...((((((((.....))))))))))))))............ ( -30.34) >DroYak_CAF1 40240 102 + 1 UUUGUUUGGUAAUGACUGUAUGAUGAGUUUGAGUCGUUGUCCUUGCCGAAGUGGUUGGUGUAACUGAGGACUUUUGGUCUUCAGAGACAAUAUCAACGAUUU ....((((((((.(((...(((((........))))).))).))))))))((.((((((((..((((((((.....)))))))).....)))))))).)).. ( -33.60) >consensus UUUGCUUGGUAAUGACAGUAUGAUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUU .(((.(((((((((((................)))))((((..((((((.....))))))...((((((((.....)))))))).)))).)))))))))... (-26.31 = -25.95 + -0.36)

| Location | 858,081 – 858,183 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 96.27 |
| Mean single sequence MFE | -19.71 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 858081 102 - 1281640 AAAUCGUUGAUAUUGUCUCUGAAGACCAGGAGUCGUCAGUUACGCCAACCACUUCGGCGAGGACAACAACUCAAACUCAUCAUACUGUCAUUACCAAGCAAA .....((((...((((((((((.(((.....))).))))...((((.........)))).))))))))))................................ ( -20.00) >DroSec_CAF1 38304 102 - 1 AAAUCGUUGAUAUUGUCUCUGAAGACCAGGAGUCCUCAGUUACGCCAACCACUUCGGCGAGGACAACAACUCAAACUCAUCAUACUGUCAUUACCAAGCAAA .....((((...((((((((((.(((.....))).))))...((((.........)))).))))))))))................................ ( -20.00) >DroSim_CAF1 33399 102 - 1 AAAUCGUUGAUAUUGUCUCUGAAGACCAGGAGUCCUCAGUUACGCCAACCACUUCGGCGAGGACAACAACUCAAACUCAUCAUACUGUCAUUACCAAGCAAA .....((((...((((((((((.(((.....))).))))...((((.........)))).))))))))))................................ ( -20.00) >DroEre_CAF1 38116 102 - 1 AAAUCGUUGAUAUUGUCUCUGAAGACCAGGAGUCCUCAGUUACGCCAACCACUUCGGCGAGGACAACGACUCAAACUCAUCAUACAGUCAUUACCAAACAAA .....(((....((((((((((.(((.....))).))))...((((.........)))).)))))).((((..............)))).......)))... ( -20.24) >DroYak_CAF1 40240 102 - 1 AAAUCGUUGAUAUUGUCUCUGAAGACCAAAAGUCCUCAGUUACACCAACCACUUCGGCAAGGACAACGACUCAAACUCAUCAUACAGUCAUUACCAAACAAA ...((((((....(((..((((.(((.....))).))))..)))((..((.....))...)).))))))................................. ( -18.30) >consensus AAAUCGUUGAUAUUGUCUCUGAAGACCAGGAGUCCUCAGUUACGCCAACCACUUCGGCGAGGACAACAACUCAAACUCAUCAUACUGUCAUUACCAAGCAAA .....((((...((((((((((.(((.....))).))))...((((.........)))).))))))))))................................ (-18.14 = -18.30 + 0.16)

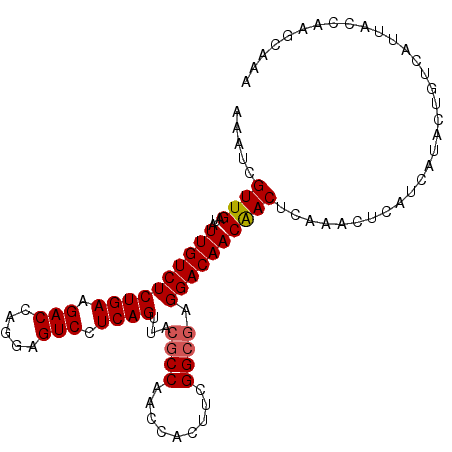
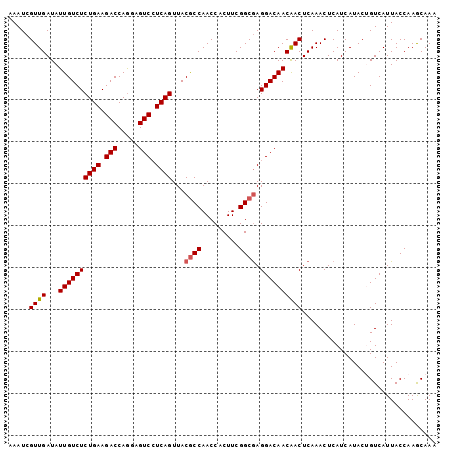
| Location | 858,103 – 858,223 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.61 |
| Mean single sequence MFE | -40.45 |
| Consensus MFE | -29.31 |
| Energy contribution | -31.23 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 858103 120 + 1281640 AUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGACGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCG .((((((..((((((((.....(((((.....)))))((..((((.(((.....))).))))..)).....))))))))..))).)))...((((((.(((......)))...)))))). ( -41.90) >DroGri_CAF1 67216 90 + 1 CUGUGUCUGUGUGGCAACC-U--------------------------GCCCAUCAUCUUC---CACAAUGUCCACAAUUUGCACAUCGGGCGCCAGCAGCGAGGCAACAUCAAAUUGCCU ..(((((((((((.(((..-(--------------------------(.....(((....---....)))....))..))))))).)))))))........((((((.......)))))) ( -20.90) >DroSec_CAF1 38326 120 + 1 AUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACGUCGGGUGCCAGUAGCGAAGCCACAUCAAACUGGCG .((((((..((((((((.....(((((.....)))))((..((((((((.....))))))))..)).....))))))))..))).)))...((((((...((.......))..)))))). ( -43.00) >DroSim_CAF1 33421 120 + 1 AUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACGUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCG .((((((..((((((((.....(((((.....)))))((..((((((((.....))))))))..)).....))))))))..))).)))...((((((.(((......)))...)))))). ( -45.70) >DroEre_CAF1 38138 120 + 1 AUGAGUUUGAGUCGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCG .((((((..((((((((.....(((((.....)))))((..((((((((.....))))))))..)).....))))))))..))).)))...((((((.(((......)))...)))))). ( -47.80) >DroYak_CAF1 40262 120 + 1 AUGAGUUUGAGUCGUUGUCCUUGCCGAAGUGGUUGGUGUAACUGAGGACUUUUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGUGAAGCCACGUCAAAUUGGCG .((((((..((((((((.((..(((.....))).))(((..((((((((.....))))))))..)))....))))))))..))).)))...((((((.(((....))).....)))))). ( -43.40) >consensus AUGAGUUUGAGUUGUUGUCCUCGCCGAAGUGGUUGGCGUAACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCG ....(((..((((((((....((((((.....))))))...((((((((.....)))))))).........))))))))..))).......((((((...((.......))..)))))). (-29.31 = -31.23 + 1.92)

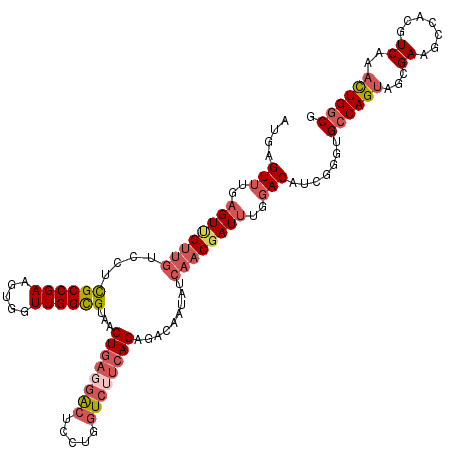
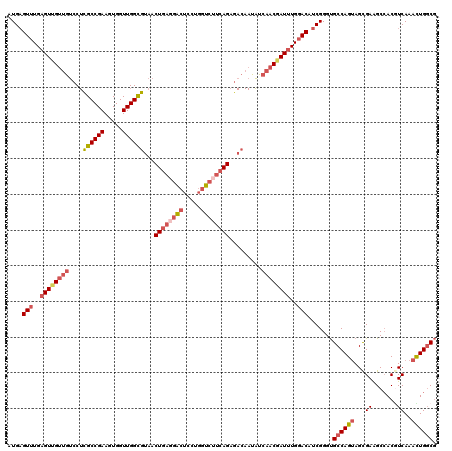
| Location | 858,143 – 858,263 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.28 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -34.84 |
| Energy contribution | -36.82 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 858143 120 + 1281640 ACUGACGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCGCUUCCGCGUCUGCGAAAACAUAUUCGCGAUUGUUGUUGUC .((((.(((.....))).)))).(((((...((((((((.((((..(((((((((((.(((......)))...))))))))..))).))))(((((......)))))))))))))))))) ( -44.60) >DroGri_CAF1 67236 110 + 1 -------GCCCAUCAUCUUC---CACAAUGUCCACAAUUUGCACAUCGGGCGCCAGCAGCGAGGCAACAUCAAAUUGCCUUUUGCGCGUCUGCGAAUAAAGAUUCGCAAUUGUUGUCGUC -------.............---....(((.(.((((((........((((((..((((.(((((((.......)))))))))))))))))((((((....)))))))))))).).))). ( -31.40) >DroSec_CAF1 38366 120 + 1 ACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACGUCGGGUGCCAGUAGCGAAGCCACAUCAAACUGGCGCUUUCGCGUCUGCGAAAACAUAUUCGCGAUAGUUGUUGUC .((((((((.....)))))))).(((((...((((.....((((((.((((((((((...((.......))..))))))))))..))))))(((((......)))))....))))))))) ( -46.00) >DroSim_CAF1 33461 120 + 1 ACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACGUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCGCUUUCGCGUCUGCGAAAACAUAUUCGCGAUAGUUGUUGUC .((((((((.....)))))))).(((((...((((.....((((((.((((((((((.(((......)))...))))))))))..))))))(((((......)))))....))))))))) ( -48.70) >DroEre_CAF1 38178 120 + 1 ACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCGCUUCCGUGUCUGCGAAAACAUAUUCGCGAUUGUUGUUGUC .((((((((.....)))))))).(((((...((((((((.(((((.(((((((((((.(((......)))...))))))))..))))))))(((((......)))))))))))))))))) ( -51.10) >DroYak_CAF1 40302 120 + 1 ACUGAGGACUUUUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGUGAAGCCACGUCAAAUUGGCGCUUCCGUGUCUGCGAAAACAUAUUCGCGAUUGUUGUUGUC .((((((((.....)))))))).(((((...((((((((.(((((.(((((((((((.(((....))).....))))))))..))))))))(((((......)))))))))))))))))) ( -48.90) >consensus ACUGAGGACUCCUGGUCUUCAGAGACAAUAUCAACGAUUUGGACAUCGGGUGCCAGUAGCGAAGCCACGUCAAACUGGCGCUUCCGCGUCUGCGAAAACAUAUUCGCGAUUGUUGUUGUC .((((((((.....)))))))).(((((...((((((((.((((((.((((((((((...((.......))..))))))))))..))))))(((((......)))))))))))))))))) (-34.84 = -36.82 + 1.97)

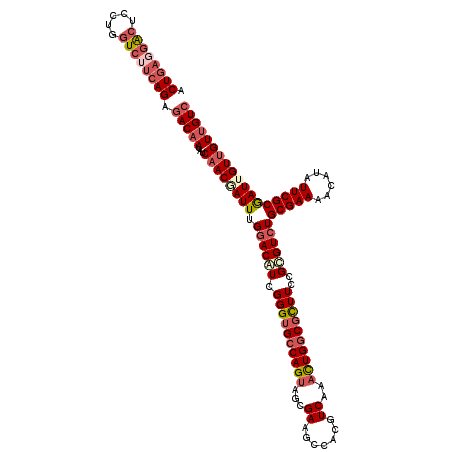
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:02 2006