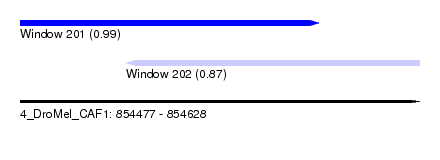
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 854,477 – 854,628 |
| Length | 151 |
| Max. P | 0.985094 |

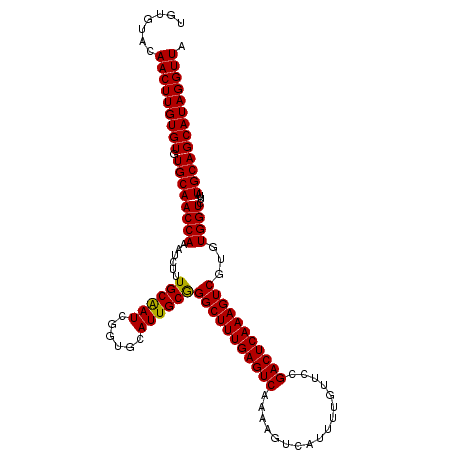
| Location | 854,477 – 854,590 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -36.57 |
| Energy contribution | -36.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 854477 113 + 1281640 UGUGUACAACUUGUGUGUGCAACCAAAUCUUUGCAAUCGGUGCAUUGCAGGCUUUGAGUCAAAAGUCAUUCUGUUCCGACUCAAAGUCGUGUGGUCUUAUGCAGCAUAGGUUA .......(((((((((.((((((((......((((((......))))))(((((((((((...((.....)).....)))))))))))...))))....))))))))))))). ( -37.50) >DroSec_CAF1 34559 113 + 1 UGUGUACAACUUGUGUGUGCAACCAAAUCUUUGCAAUCGGUGCAUUGCGGGCUUUGAGUCAAAAGUCAUUUUGUUCCGACUCAAAGUCGUGUGGUCUUAUGCAGCAUAGGUUA .......(((((((((.((((((((......((((((......))))))(((((((((((.................)))))))))))...))))....))))))))))))). ( -35.93) >DroSim_CAF1 29349 113 + 1 UGUGUACAACUUGUGUGUGCAACCAAAUCUUUGCGAUCGGUGCAUUGCGGGCUUUGAGUCAAAAGUCAUUUUGUUCCGACUCAAAGUCGUUUGGUCUUAUGCAGCAUAGGUUA .......(((((((((.((((((((((((((((.(.((((.(((.....((((((......))))))....))).))))).)))))..)))))))....))))))))))))). ( -38.70) >consensus UGUGUACAACUUGUGUGUGCAACCAAAUCUUUGCAAUCGGUGCAUUGCGGGCUUUGAGUCAAAAGUCAUUUUGUUCCGACUCAAAGUCGUGUGGUCUUAUGCAGCAUAGGUUA .......(((((((((.((((((((......((((((......))))))(((((((((((.................)))))))))))...))))....))))))))))))). (-36.57 = -36.13 + -0.44)

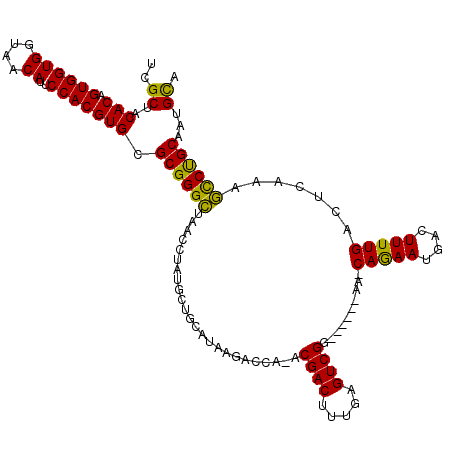
| Location | 854,517 – 854,628 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -26.86 |
| Energy contribution | -25.66 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 854517 111 - 1281640 UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGCUAACCUAUGCUGCAUAAGACCACACGACUUUGAGUCGG------AA-CAGAAUGACUUUUGACUCAAAGCCUGCAAUGCA ..((..(((.((((((....))..))))))).(((((((..(.((((...)))).)...........(((((((((------((-.........))))))))))))))))))...)). ( -32.40) >DroSec_CAF1 34599 111 - 1 UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGCUAACCUAUGCUGCAUAAGACCACACGACUUUGAGUCGG------AA-CAAAAUGACUUUUGACUCAAAGCCCGCAAUGCA ..((..(((.((((((....))..))))))).(((((((..(.((((...)))).)...........(((((((((------((-.........))))))))))))))))))...)). ( -34.60) >DroSim_CAF1 29389 111 - 1 UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGCUAACCUAUGCUGCAUAAGACCAAACGACUUUGAGUCGG------AA-CAAAAUGACUUUUGACUCAAAGCCCGCAAUGCA ..((..(((.((((((....))..))))))).(((((((..(.((((...)))).)...........(((((((((------((-.........))))))))))))))))))...)). ( -34.60) >DroEre_CAF1 34980 97 - 1 UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGCUAACUUAUGC--------------CGACUUUGGGUCGG------AAUCAGAAUGACUUUUGACUCAA-GCCUGCAAUGUA (((((((...))))))).........((((..(((((((........(--------------((((.....)))))------..((((((....))))))....)-)))))).)))). ( -30.90) >DroYak_CAF1 36929 103 - 1 UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGUAAACUUAUGC--------------CGACUUUGAGUCGAAAACAAAAUCAGAAUGACUUUUGACUCAA-AUCUGCAAUGUA ((((..(((.((((((....))..))))))).)))).........(((--------------.((.((((((((((((................)))))))))))-))).)))..... ( -27.59) >consensus UCGCUACACAGUGGUGGUAACAUUCCACGUGCGCGGGCUAACCUAUGCUGCAUAAGACCA_ACGACUUUGAGUCGG______AA_CAGAAUGACUUUUGACUCAAAGCCUGCAAUGCA ..((..(((.((((((....))..))))))).((((((........................((((.....))))..........(((((....))))).......))))))...)). (-26.86 = -25.66 + -1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:56 2006