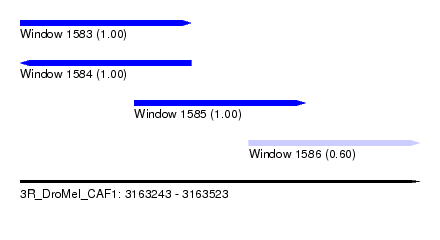
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,163,243 – 3,163,523 |
| Length | 280 |
| Max. P | 0.999918 |

| Location | 3,163,243 – 3,163,363 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -45.00 |
| Energy contribution | -44.64 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3163243 120 + 27905053 UUUGUGCCCCGGUGGGCGUUGCCCAGAGCGAAAAUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGU .....((((....)))).((((.(((..((((((((((..(((((.((((..((((((...))))))....))))...)))))(((((.....))))).)))))))))).))).)))).. ( -47.20) >DroSec_CAF1 11134 120 + 1 UUUGUGCCCCGGUGGGCGUUGCCCAGAGCGAAAAUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGU .....((((....)))).((((.(((..((((((((((..(((((.((((..((((((...))))))....))))...)))))(((((.....))))).)))))))))).))).)))).. ( -47.20) >DroSim_CAF1 13303 120 + 1 UUUGCGCCCCGGUGGGCGUUGCCCAGAGCGAAAAUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGU ...((((((....))))))(((.(((..((((((((((..(((((.((((..((((((...))))))....))))...)))))(((((.....))))).)))))))))).))).)))... ( -49.30) >DroEre_CAF1 2812 119 + 1 UUUGUGGCC-GGUGGGCGUUGCCCAGAGCGAAAGUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGAUGGAGGAGAAGUUUUCGGCUGCGCAAGU ...(..(((-((..((((((((.(..(((....)))..)..(((....)))...))))))))..)..........((((.((.(((((.....))))).)).)))).))))..)...... ( -45.80) >DroYak_CAF1 12532 120 + 1 UUUGUGUCCUGGUGGGCGUUGCCCAGAGUGAACAUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGACGGAGGAGAAAUUUUCGGCUGCGCAAGU .....((((....)))).((((.(((..((((.(((((..(((((.((((..((((((...))))))....))))...)))))(((((.....))))).))))).)))).))).)))).. ( -40.50) >consensus UUUGUGCCCCGGUGGGCGUUGCCCAGAGCGAAAAUUUCGAUGACGAGUGUCCUUGCAGCGUCUGCAAAUCUGCAUAGACGUCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGU (((((((.....(((((...))))).((((((((((((..(((((.((((..((((((...))))))....))))...)))))(((((.....))))).))))))))).)))))))))). (-45.00 = -44.64 + -0.36)

| Location | 3,163,243 – 3,163,363 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -44.56 |
| Consensus MFE | -42.10 |
| Energy contribution | -43.30 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.55 |
| SVM RNA-class probability | 0.999918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3163243 120 - 27905053 ACUUGCGCAGCCGAAAAUUUCUCCUCCAUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAAUUUUCGCUCUGGGCAACGCCCACCGGGGCACAAA ..((((.(((.((((((((((.(((((.....)))))((((((((.(((((...........))))).))))....))))..))))))))))..))).)))).((((....))))..... ( -48.10) >DroSec_CAF1 11134 120 - 1 ACUUGCGCAGCCGAAAAUUUCUCCUCCAUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAAUUUUCGCUCUGGGCAACGCCCACCGGGGCACAAA ..((((.(((.((((((((((.(((((.....)))))((((((((.(((((...........))))).))))....))))..))))))))))..))).)))).((((....))))..... ( -48.10) >DroSim_CAF1 13303 120 - 1 ACUUGCGCAGCCGAAAAUUUCUCCUCCAUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAAUUUUCGCUCUGGGCAACGCCCACCGGGGCGCAAA ..((((.(((.((((((((((.(((((.....)))))((((((((.(((((...........))))).))))....))))..))))))))))..))).))))(((((....))))).... ( -49.40) >DroEre_CAF1 2812 119 - 1 ACUUGCGCAGCCGAAAACUUCUCCUCCAUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAACUUUCGCUCUGGGCAACGCCCACC-GGCCACAAA ..((((.(((.(((((..(((.(((((.....)))))((((((((.(((((...........))))).))))....))))..)))..)))))..))).)))).(((....-)))...... ( -38.00) >DroYak_CAF1 12532 120 - 1 ACUUGCGCAGCCGAAAAUUUCUCCUCCGUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAAUGUUCACUCUGGGCAACGCCCACCAGGACACAAA .((((.(((((........((.(((((.....))))).)).((((..((......)))))))))))..(((((.(((....)))..)))))....(((((...))))).))))....... ( -39.20) >consensus ACUUGCGCAGCCGAAAAUUUCUCCUCCAUCAGGGAGGUGACGUCUAUGCAGAUUUGCAGACGCUGCAAGGACACUCGUCAUCGAAAUUUUCGCUCUGGGCAACGCCCACCGGGGCACAAA ..((((.(((.((((((((((.(((((.....)))))((((((((.(((((...........))))).))))....))))..))))))))))..))).)))).((((....))))..... (-42.10 = -43.30 + 1.20)

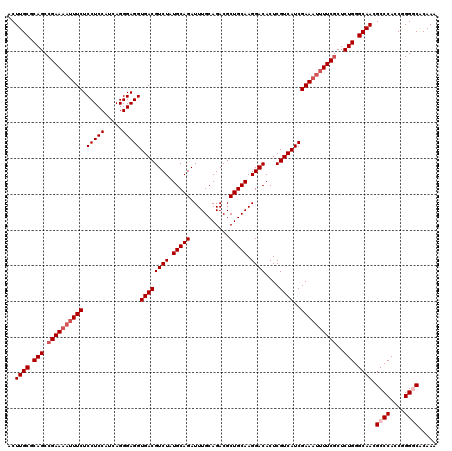
| Location | 3,163,323 – 3,163,443 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -35.94 |
| Energy contribution | -35.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3163323 120 + 27905053 UCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGUCUCUAAAAAGGGGAGCCACGCCCCUAGCACUAGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAGC ((.(((((.....))))).)).......((((((....))((((....)))).)))).((((((..(((....)))..............((((.....))))))))))........... ( -38.50) >DroSec_CAF1 11214 120 + 1 UCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGUCUCUAAAAAGGGCAGCCACGCCCCUAACACCGGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAAC ((.(((((.....))))).)).......((((((.(.............).)))))).((((((.........(((..........))).((((.....))))))))))........... ( -38.22) >DroSim_CAF1 13383 120 + 1 UCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGUCUCUAAAAAGGGGAGCCACGCCCCUAGCUCUGGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAAC ((.(((((.....))))).)).......((((((....))((((....)))).)))).((((((..((......))..............((((.....))))))))))........... ( -37.10) >DroEre_CAF1 2891 120 + 1 UCACCUCCCUGAUGGAGGAGAAGUUUUCGGCUGCGCAAGUCUCUAAAAAGGGGAGCCACGCCCCUCGCAGCGGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAAC ((.(((((.....))))).)).......((((((....))((((....)))).)))).((((((..(((....)))..............((((.....))))))))))........... ( -37.30) >DroYak_CAF1 12612 120 + 1 UCACCUCCCUGACGGAGGAGAAAUUUUCGGCUGCGCAAGUCUCUAUAAAGGGGAGCCACGCCCCCAGCACUGGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAAC ((.(((((.....))))).)).......((((((....))((((....)))).)))).((((((..(((....)))..............((((.....))))))))))........... ( -38.20) >consensus UCACCUCCCUGAUGGAGGAGAAAUUUUCGGCUGCGCAAGUCUCUAAAAAGGGGAGCCACGCCCCUAGCACUGGUGCAACAAAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAAC ((.(((((.....))))).)).......((((((....))((((....)))).)))).((((((.........(((..........))).((((.....))))))))))........... (-35.94 = -35.94 + 0.00)

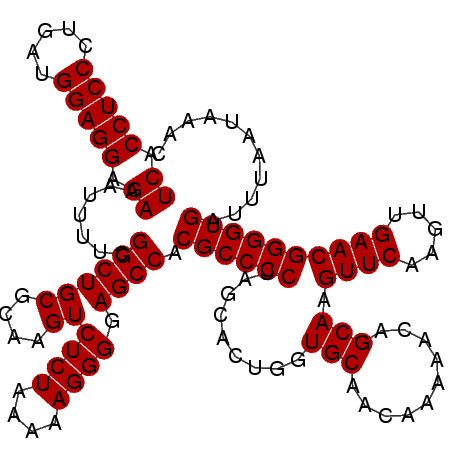

| Location | 3,163,403 – 3,163,523 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -26.75 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.84 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3163403 120 + 27905053 AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAGCGGACUUGCAAAAAAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGCAAACAACAAUGUUGACAUUGAUAUUGCCGUGUG ......((((((((..((((((........))))))....))))))))......((((((........(((((.((((((...(((((........)))))))))))))))).)))))). ( -28.90) >DroSec_CAF1 11294 120 + 1 AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAACGGACUUGCAAAAAAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGCAAACAACAAAGUUGACAUUGAUAUCGCCGUGUG ......((((((((..((((((........))))))....))))))))......((((((........(((((((.((....)).))...((((...))))))))).......)))))). ( -27.06) >DroSim_CAF1 13463 120 + 1 AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAACGGACUUGCAAAAAAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGCAAACAACAAAGUUGACAUUGAUAUCGCCGUGUG ......((((((((..((((((........))))))....))))))))......((((((........(((((((.((....)).))...((((...))))))))).......)))))). ( -27.06) >DroEre_CAF1 2971 117 + 1 AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAACGGACUGGCGAAAAAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGCAAGCAACAAAGUUGACAUUGAUAUCG---UGCG ......((.(((((..((((((........))))))....))))).))......((((((..((((......(((.((....)).))).(((((...)))).).))))..)))---))). ( -21.10) >DroYak_CAF1 12692 120 + 1 AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAACGGACUUGCGAAACAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGGCAACAACAAAGUUGACAUUGAUAUCGCCCUGUG ....((((........)))).((((((((((.........(..(.(((......))).)..)......(((((.((((.......(....)......)))))))))...)))))))))). ( -29.62) >consensus AAAACAGCAAGUUCAAGUUGAACGGGGUGAUUUAAUAAACGGACUUGCAAAAAAGCACGGACUCGGAAAAUGUGCAAUGUAAAUAGCAAACAACAAAGUUGACAUUGAUAUCGCCGUGUG ......((((((((..((((((........))))))....))))))))......((((((........(((((((.((....)).))...((((...))))))))).......)))))). (-21.80 = -22.84 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:28 2006